2WJU
 
 | |
1QIP
 
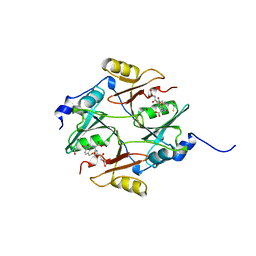 | | HUMAN GLYOXALASE I COMPLEXED WITH S-P-NITROBENZYLOXYCARBONYLGLUTATHIONE | | Descriptor: | BETA-MERCAPTOETHANOL, PROTEIN (LACTOYLGLUTATHIONE LYASE), S-P-NITROBENZYLOXYCARBONYLGLUTATHIONE, ... | | Authors: | Cameron, A.D, Ridderstrom, M, Olin, B, Mannervik, B. | | Deposit date: | 1999-06-14 | | Release date: | 1999-11-24 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (1.72 Å) | | Cite: | Reaction mechanism of glyoxalase I explored by an X-ray crystallographic analysis of the human enzyme in complex with a transition state analogue.
Biochemistry, 38, 1999
|
|
1QH5
 
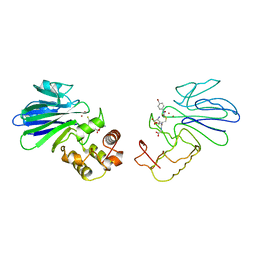 | | HUMAN GLYOXALASE II WITH S-(N-HYDROXY-N-BROMOPHENYLCARBAMOYL)GLUTATHIONE | | Descriptor: | GLUTATHIONE, PROTEIN (HYDROXYACYLGLUTATHIONE HYDROLASE), S-(N-HYDROXY-N-BROMOPHENYLCARBAMOYL)GLUTATHIONE, ... | | Authors: | Cameron, A.D, Ridderstrom, M, Olin, B, Mannervik, B. | | Deposit date: | 1999-05-11 | | Release date: | 1999-09-24 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (1.45 Å) | | Cite: | Crystal structure of human glyoxalase II and its complex with a glutathione thiolester substrate analogue.
Structure Fold.Des., 7, 1999
|
|
2WAS
 
 | |
2WAT
 
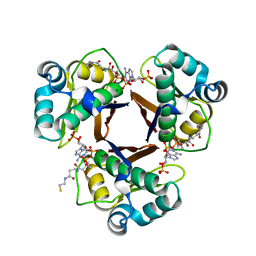 | | Structure of the fungal type I FAS PPT domain in complex with CoA | | Descriptor: | 3-OXOACYL-[ACYL-CARRIER-PROTEIN] SYNTHASE, CHLORIDE ION, COENZYME A, ... | | Authors: | Johansson, P, Mulincacci, B, Koestler, C, Grininger, M. | | Deposit date: | 2009-02-15 | | Release date: | 2009-08-25 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Multimeric Options for the Auto-Activation of the Saccharomyces Cerevisiae Fas Type I Megasynthase.
Structure, 17, 2009
|
|
1OCU
 
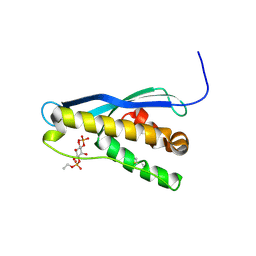 | | Crystal structure of the yeast PX-domain protein Grd19p (sorting nexin 3) complexed to phosphatidylinosytol-3-phosphate. | | Descriptor: | 2-(BUTANOYLOXY)-1-{[(HYDROXY{[2,3,4,6-TETRAHYDROXY-5-(PHOSPHONOOXY)CYCLOHEXYL]OXY}PHOSPHORYL)OXY]METHYL}ETHYL BUTANOATE, SORTING NEXIN | | Authors: | Zhou, C.Z, Li de La Sierra-Gallay, I, Cheruel, S, Collinet, B, Minard, P, Blondeau, K, Henkes, G, Aufrere, R, Leulliot, N, Graille, M, Sorel, I, Savarin, P, de la Torre, F, Poupon, A, Janin, J, van Tilbeurgh, H. | | Deposit date: | 2003-02-10 | | Release date: | 2003-12-12 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Crystal structure of the yeast Phox homology (PX) domain protein Grd19p complexed to phosphatidylinositol-3-phosphate.
J. Biol. Chem., 278, 2003
|
|
2WB7
 
 | | pT26-6p | | Descriptor: | PT26-6P | | Authors: | Keller, J, Leulliot, N, Soler, N, Collinet, B, Vincentelli, R, Forterre, P, van Tilbeurgh, H. | | Deposit date: | 2009-02-22 | | Release date: | 2009-03-03 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | A Protein Encoded by a New Family of Mobile Elements from Euryarchaea Exhibits Three Domains with Novel Folds.
Protein Sci., 18, 2009
|
|
1OCS
 
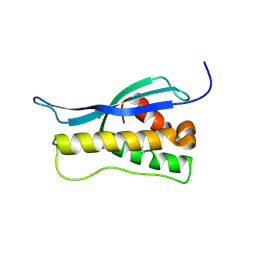 | | Crystal structure of the yeast PX-doamin protein Grd19p (sorting nexin3) complexed to phosphatidylinosytol-3-phosphate. | | Descriptor: | GLYCEROL, SORTING NEXIN GRD19 | | Authors: | Zhou, C.Z, Li De La Sierra-Gallay, I, Cheruel, S, Collinet, B, Minard, P, Blondeau, K, Henkes, G, Aufrere, R, Leulliot, N, Graille, M, Sorel, I, Savarin, P, De La Torre, F, Poupon, A, Janin, J, Van Tilbeurgh, H. | | Deposit date: | 2003-02-10 | | Release date: | 2003-12-12 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (2.03 Å) | | Cite: | Crystal Structure of the Yeast Phox Homology (Px) Protein Grd19P (Sorting Nexin 3) Complexed to Phosphatidylinositol-3-Phosphate
J.Biol.Chem., 278, 2003
|
|
2WB6
 
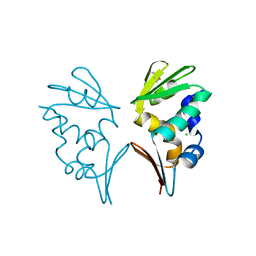 | | Crystal structure of AFV1-102, a protein from the Acidianus Filamentous Virus 1 | | Descriptor: | AFV1-102, CHLORIDE ION | | Authors: | Keller, J, Leulliot, N, Collinet, B, Campanacci, V, Cambillau, C, Pranghisvilli, D, van Tilbeurgh, H. | | Deposit date: | 2009-02-22 | | Release date: | 2009-03-03 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | Crystal Structure of Afv1-102, a Protein from the Acidianus Filamentous Virus 1.
Protein Sci., 18, 2009
|
|
2VWB
 
 | | Structure of the archaeal Kae1-Bud32 fusion protein MJ1130: a model for the eukaryotic EKC-KEOPS subcomplex involved in transcription and telomere homeostasis. | | Descriptor: | PHOSPHOAMINOPHOSPHONIC ACID-ADENYLATE ESTER, PUTATIVE O-SIALOGLYCOPROTEIN ENDOPEPTIDASE | | Authors: | Hecker, A, Lopreiato, R, Graille, M, Collinet, B, Forterre, P, Domenico, L, van Tilbeurgh, H. | | Deposit date: | 2008-06-20 | | Release date: | 2008-08-19 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (3.05 Å) | | Cite: | Structure of the Archaeal Kae1/Bud32 Fusion Protein Mj1130: A Model for the Eukaryotic Ekc/Keops Subcomplex
Embo J., 27, 2008
|
|
1QFG
 
 | | E. COLI FERRIC HYDROXAMATE RECEPTOR (FHUA) | | Descriptor: | 3-HYDROXY-TETRADECANOIC ACID, DECYLAMINE-N,N-DIMETHYL-N-OXIDE, DIPHOSPHATE, ... | | Authors: | Ferguson, A.D, Welte, W, Hofmann, E, Lindner, B, Holst, O, Coulton, J.W, Diederichs, K. | | Deposit date: | 1999-04-10 | | Release date: | 2000-07-26 | | Last modified: | 2022-12-21 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | A conserved structural motif for lipopolysaccharide recognition by procaryotic and eucaryotic proteins.
Structure Fold.Des., 8, 2000
|
|
6N5X
 
 | | Crystal structure of the SNX5 PX domain in complex with the CI-MPR (space group P212121 - Form 1) | | Descriptor: | GLYCEROL, O-(O-(2-AMINOPROPYL)-O'-(2-METHOXYETHYL)POLYPROPYLENE GLYCOL 500), Sorting nexin-5,Cation-independent mannose-6-phosphate receptor | | Authors: | Collins, B, Paul, B, Weeratunga, S. | | Deposit date: | 2018-11-22 | | Release date: | 2019-09-18 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (2.051 Å) | | Cite: | Molecular identification of a BAR domain-containing coat complex for endosomal recycling of transmembrane proteins.
Nat.Cell Biol., 21, 2019
|
|
6N5Z
 
 | |
6N5Y
 
 | |
4QDH
 
 | |
3LH9
 
 | |
3LH8
 
 | |
3LHA
 
 | |
8CMX
 
 | |
6C3L
 
 | | Crystal structure of BCL6 BTB domain with compound 15f | | Descriptor: | B-cell lymphoma 6 protein, N-[2-(1H-indol-3-yl)ethyl]-N'-{3-[(4-methylpiperazin-1-yl)methyl]-1-[2-(morpholin-4-yl)-2-oxoethyl]-1H-indol-6-yl}thiourea | | Authors: | Linhares, B, Cheng, H, Cierpicki, T, Xue, F. | | Deposit date: | 2018-01-10 | | Release date: | 2019-01-16 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (1.46092153 Å) | | Cite: | Identification of Thiourea-Based Inhibitors of the B-Cell Lymphoma 6 BTB Domain via NMR-Based Fragment Screening and Computer-Aided Drug Design.
J.Med.Chem., 61, 2018
|
|
6C3N
 
 | | Crystal structure of BCL6 BTB domain in complex with compound 7CC5 | | Descriptor: | B-cell lymphoma 6 protein, N-(2-phenylethyl)-N'-pyridin-3-ylthiourea | | Authors: | Linhares, B, Cheng, H, Xue, F, Cierpicki, T. | | Deposit date: | 2018-01-10 | | Release date: | 2019-01-16 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (2.53170586 Å) | | Cite: | Identification of Thiourea-Based Inhibitors of the B-Cell Lymphoma 6 BTB Domain via NMR-Based Fragment Screening and Computer-Aided Drug Design.
J.Med.Chem., 61, 2018
|
|
5TGH
 
 | |
6VRW
 
 | | Cryo-EM structure of stabilized HIV-1 Env trimer CAP256.wk34.c80 SOSIP.RnS2 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Envelope glycoprotein gp120, ... | | Authors: | Gorman, J, Kwong, P.D. | | Deposit date: | 2020-02-10 | | Release date: | 2020-03-11 | | Last modified: | 2021-10-06 | | Method: | ELECTRON MICROSCOPY (3.71 Å) | | Cite: | Structure of Super-Potent Antibody CAP256-VRC26.25 in Complex with HIV-1 Envelope Reveals a Combined Mode of Trimer-Apex Recognition.
Cell Rep, 31, 2020
|
|
5XVA
 
 | | Crystal Structure of PAK4 in complex with inhibitor CZH216 | | Descriptor: | ETHANOL, Serine/threonine-protein kinase PAK 4, [6-chloranyl-4-[(5-methyl-1H-pyrazol-3-yl)amino]quinazolin-2-yl]-[(3R)-3-methylpiperazin-1-yl]methanone | | Authors: | Zhao, F, Li, H. | | Deposit date: | 2017-06-27 | | Release date: | 2018-02-07 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (1.847 Å) | | Cite: | Structure-Based Design of 6-Chloro-4-aminoquinazoline-2-carboxamide Derivatives as Potent and Selective p21-Activated Kinase 4 (PAK4) Inhibitors.
J. Med. Chem., 61, 2018
|
|
5XVG
 
 | | Crystal Structure of PAK4 in complex with inhibitor CZH226 | | Descriptor: | 1,2-ETHANEDIOL, ETHANOL, Serine/threonine-protein kinase PAK 4, ... | | Authors: | Zhao, F, Li, H. | | Deposit date: | 2017-06-27 | | Release date: | 2018-02-07 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Structure-Based Design of 6-Chloro-4-aminoquinazoline-2-carboxamide Derivatives as Potent and Selective p21-Activated Kinase 4 (PAK4) Inhibitors.
J. Med. Chem., 61, 2018
|
|
