1AY7
 
 | | RIBONUCLEASE SA COMPLEX WITH BARSTAR | | Descriptor: | BARSTAR, GUANYL-SPECIFIC RIBONUCLEASE SA | | Authors: | Sevcik, J, Urbanikova, L, Dauter, Z, Wilson, K.S. | | Deposit date: | 1997-11-14 | | Release date: | 1999-03-02 | | Last modified: | 2023-08-02 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Recognition of RNase Sa by the inhibitor barstar: structure of the complex at 1.7 A resolution.
Acta Crystallogr.,Sect.D, 54, 1998
|
|
1BWW
 
 | | BENCE-JONES IMMUNOGLOBULIN REI VARIABLE PORTION, T39K MUTANT | | Descriptor: | PROTEIN (IG KAPPA CHAIN V-I REGION REI) | | Authors: | Uson, I, Pohl, E, Schneider, T.R, Dauter, Z, Schmidt, A, Fritz, H.J, Sheldrick, G.M. | | Deposit date: | 1998-09-29 | | Release date: | 1998-10-07 | | Last modified: | 2023-08-09 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | 1.7 A structure of the stabilized REIv mutant T39K. Application of local NCS restraints.
Acta Crystallogr.,Sect.D, 55, 1999
|
|
1KTJ
 
 | | X-ray Structure Of Der P 2, The Major House Dust Mite Allergen | | Descriptor: | ALLERGEN DER P 2 | | Authors: | Derewenda, U, Li, J, Derewenda, Z, Dauter, Z, Mueller, G.A, Rule, G.S, Benjamin, D.C. | | Deposit date: | 2002-01-16 | | Release date: | 2002-05-15 | | Last modified: | 2021-10-27 | | Method: | X-RAY DIFFRACTION (2.15 Å) | | Cite: | The crystal structure of a major dust mite allergen Der p 2, and its biological implications.
J.Mol.Biol., 318, 2002
|
|
1BOX
 
 | | N39S MUTANT OF RNASE SA FROM STREPTOMYCES AUREOFACIENS | | Descriptor: | GUANYL-SPECIFIC RIBONUCLEASE SA | | Authors: | Hebert, E.J, Giletto, A, Sevcik, J, Urbanikova, L, Wilson, K.S, Dauter, Z, Pace, C.N. | | Deposit date: | 1998-08-07 | | Release date: | 1999-12-29 | | Last modified: | 2023-08-09 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Contribution of a conserved asparagine to the conformational stability of ribonucleases Sa, Ba, and T1.
Biochemistry, 37, 1998
|
|
1C5E
 
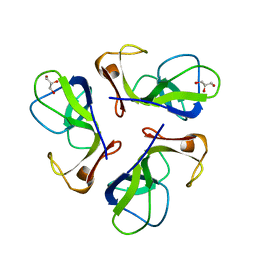 | | BACTERIOPHAGE LAMBDA HEAD PROTEIN D | | Descriptor: | GLYCEROL, HEAD DECORATION PROTEIN | | Authors: | Yang, F, Forrer, P, Dauter, Z, Pluckthun, A, Wlodawer, A. | | Deposit date: | 1999-11-18 | | Release date: | 2000-03-08 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (1.1 Å) | | Cite: | Novel fold and capsid-binding properties of the lambda-phage display platform protein gpD.
Nat.Struct.Biol., 7, 2000
|
|
1CC0
 
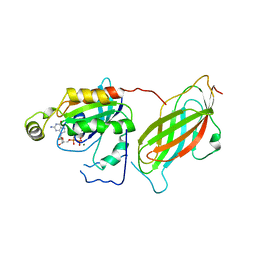 | | CRYSTAL STRUCTURE OF THE RHOA.GDP-RHOGDI COMPLEX | | Descriptor: | GUANOSINE-5'-DIPHOSPHATE, MAGNESIUM ION, rho GDP dissociation inhibitor alpha, ... | | Authors: | Longenecker, K.L, Read, P, Derewenda, U, Dauter, Z, Garrard, S, Walker, L, Somlyo, A.V, Somlyo, A.P, Nakamoto, R.K, Derewenda, Z.S. | | Deposit date: | 1999-03-03 | | Release date: | 2000-01-07 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (5 Å) | | Cite: | How RhoGDI binds Rho.
Acta Crystallogr.,Sect.D, 55, 1999
|
|
1E6F
 
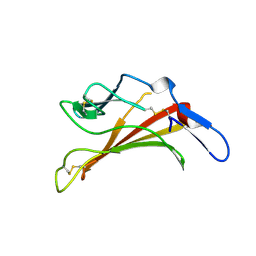 | | Human MIR-receptor, repeat 11 | | Descriptor: | CATION-INDEPENDENT MANNOSE-6-PHOSPHATE RECEPTOR | | Authors: | Von Buelow, R, Rajashankar, K.R, Dauter, M, Dauter, Z, Grimme, S, Schmidt, B, Von Figura, K, Uson, I. | | Deposit date: | 2000-08-15 | | Release date: | 2001-08-09 | | Last modified: | 2019-07-24 | | Method: | X-RAY DIFFRACTION (1.75 Å) | | Cite: | Locating the Anomalous Scatterer Substructures in Halide and Sulfur Phasing
Acta Crystallogr.,Sect.D, 59, 2003
|
|
6B91
 
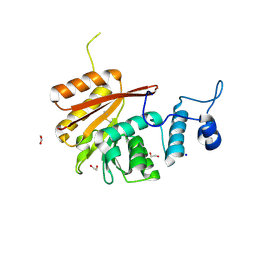 | | Crystal structure of the N-terminal domain of human METTL16 | | Descriptor: | 1,2-ETHANEDIOL, SODIUM ION, U6 small nuclear RNA (adenine-(43)-N(6))-methyltransferase | | Authors: | Ruszkowska, A, Ruszkowski, M, Dauter, Z, Brown, J.A. | | Deposit date: | 2017-10-09 | | Release date: | 2018-04-04 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (1.94 Å) | | Cite: | Structural insights into the RNA methyltransferase domain of METTL16.
Sci Rep, 8, 2018
|
|
3PVA
 
 | | PENICILLIN V ACYLASE FROM B. SPHAERICUS | | Descriptor: | PROTEIN (PENICILLIN V ACYLASE) | | Authors: | Suresh, C.G, Pundle, A.V, Rao, K.N, Sivaraman, H, Brannigan, J.A, Mcvey, C.E, Verma, C.S, Dauter, Z, Dodson, E.J, Dodson, G.G. | | Deposit date: | 1998-11-13 | | Release date: | 1999-11-15 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Penicillin V acylase crystal structure reveals new Ntn-hydrolase family members.
Nat.Struct.Biol., 6, 1999
|
|
3NR6
 
 | | Crystal structure of xenotropic murine leukemia virus-related virus (XMRV) protease | | Descriptor: | PHOSPHATE ION, POTASSIUM ION, Protease p14 | | Authors: | Lubkowski, J, Li, M, Gustchina, A, Zhou, D, Dauter, Z, Wlodawer, A. | | Deposit date: | 2010-06-30 | | Release date: | 2011-02-02 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (1.97 Å) | | Cite: | Crystal structure of XMRV protease differs from the structures of other retropepsins.
Nat.Struct.Mol.Biol., 18, 2011
|
|
6B92
 
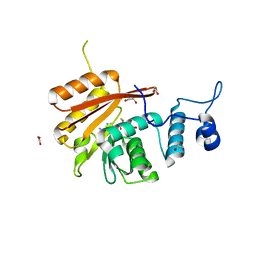 | | Crystal Structure of the N-terminal domain of human METTL16 in complex with SAH | | Descriptor: | 1,2-ETHANEDIOL, S-ADENOSYL-L-HOMOCYSTEINE, U6 small nuclear RNA (adenine-(43)-N(6))-methyltransferase | | Authors: | Ruszkowska, A, Ruszkowski, M, Dauter, Z, Brown, J.A. | | Deposit date: | 2017-10-09 | | Release date: | 2018-04-04 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Structural insights into the RNA methyltransferase domain of METTL16.
Sci Rep, 8, 2018
|
|
6BQ7
 
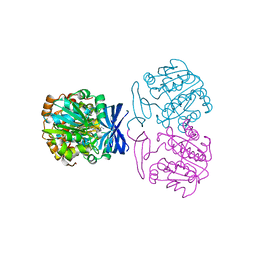 | |
6BQ2
 
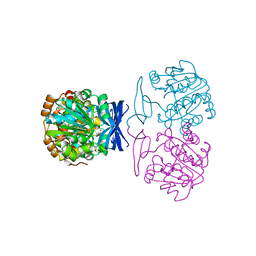 | |
1HH3
 
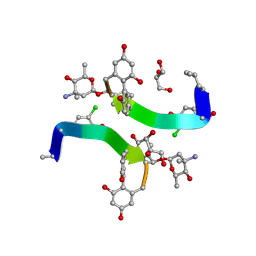 | | Decaplanin first P21-Form | | Descriptor: | 4-epi-vancosamine, DECAPLANIN, GLYCEROL, ... | | Authors: | Lehmann, C, Vertessy, L, Sheldrick, G.M, Dauter, Z, Dauter, M. | | Deposit date: | 2000-12-19 | | Release date: | 2005-07-11 | | Last modified: | 2020-07-29 | | Method: | X-RAY DIFFRACTION (1 Å) | | Cite: | Structures of Four Crystal Forms of Decaplanin
Helv.Chim.Acta, 86, 2003
|
|
1HHC
 
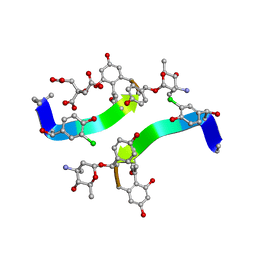 | | Crystal structure of Decaplanin - space group P21, second form | | Descriptor: | 4-epi-vancosamine, CITRIC ACID, DECAPLANIN, ... | | Authors: | Lehmann, C, Vertessy, L, Sheldrick, G.M, Dauter, Z, Dauter, M. | | Deposit date: | 2000-12-22 | | Release date: | 2005-07-11 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (1.13 Å) | | Cite: | Structures of Four Crystal Forms of Decaplanin
Helv.Chim.Acta, 86, 2003
|
|
1HHF
 
 | | Decaplanin second P6122-Form | | Descriptor: | 4-epi-vancosamine, CHLORIDE ION, DECAPLANIN, ... | | Authors: | Lehmann, C, Vertessy, L, Sheldrick, G.M, Dauter, Z, Dauter, M. | | Deposit date: | 2000-12-22 | | Release date: | 2005-07-11 | | Last modified: | 2020-07-29 | | Method: | X-RAY DIFFRACTION (1.47 Å) | | Cite: | Structures of Four Crystal Forms of Decaplanin
Helv.Chim.Acta, 86, 2003
|
|
1GQB
 
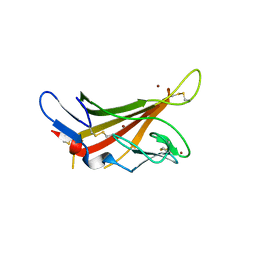 | | HUMAN MIR-RECEPTOR, REPEAT 11 | | Descriptor: | BROMIDE ION, CATION-INDEPENDENT MANNOSE-6-PHOSPHATE RECEPTOR | | Authors: | Von Buelow, R, Dauter, M, Dauter, Z, Rajashankar, K.R, Grimme, S, Schmidt, B, Von Figura, K, Uson, I. | | Deposit date: | 2001-11-22 | | Release date: | 2002-12-05 | | Last modified: | 2019-05-22 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Locating the Anomalous Scatterer Substructures in Halide and Sulfur Phasing
Acta Crystallogr.,Sect.D, 59, 2003
|
|
1SCS
 
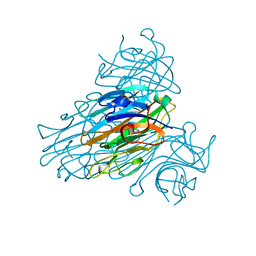 | | HIGH-RESOLUTION STRUCTURES OF SINGLE-METAL-SUBSTITUTED CONCANAVALIN A: THE CO,CA-PROTEIN AT 1.6 ANGSTROMS AND THE NI,CA-PROTEIN AT 2.0 ANGSTROMS | | Descriptor: | CALCIUM ION, COBALT (II) ION, CONCANAVALIN A | | Authors: | Emmerich, C, Helliwell, J.R, Redshaw, M, Naismith, J.H, Harrop, S.J, Raftery, J, Kalb, A.J, Yariv, J, Dauter, Z, Wilson, K.S. | | Deposit date: | 1993-12-06 | | Release date: | 1994-05-31 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | High-resolution structures of single-metal-substituted concanavalin A: the Co,Ca-protein at 1.6 A and the Ni,Ca-protein at 2.0 A.
Acta Crystallogr.,Sect.D, 50, 1994
|
|
5VER
 
 | | MOUSE KYNURENINE AMINOTRANSFERASE III, RE-REFINEMENT OF THE PDB STRUCTURE 3E2Z | | Descriptor: | 4'-DEOXY-4'-AMINOPYRIDOXAL-5'-PHOSPHATE, 4-(2-HYDROXYETHYL)-1-PIPERAZINE ETHANESULFONIC ACID, CALCIUM ION, ... | | Authors: | Wlodawer, A, Dauter, Z, Minor, W, Stanfield, R, Porebski, P, Jaskolski, M, Pozharski, E, Weichenberger, C.X, Rupp, B. | | Deposit date: | 2017-04-05 | | Release date: | 2017-11-29 | | Last modified: | 2022-04-13 | | Method: | X-RAY DIFFRACTION (2.81 Å) | | Cite: | Detect, correct, retract: How to manage incorrect structural models.
FEBS J., 285, 2018
|
|
5VLD
 
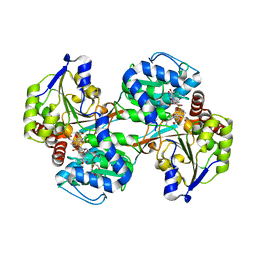 | |
1RRE
 
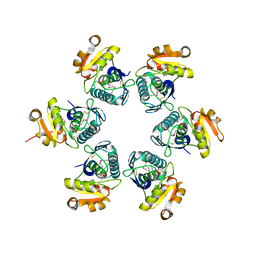 | | Crystal structure of E.coli Lon proteolytic domain | | Descriptor: | ATP-dependent protease La, SULFATE ION | | Authors: | Botos, I, Melnikov, E.E, Cherry, S, Tropea, J.E, Khalatova, A.G, Rasulova, F, Dauter, Z, Maurizi, M.R, Rotanova, T.V, Wlodawer, A, Gustchina, A. | | Deposit date: | 2003-12-08 | | Release date: | 2004-02-03 | | Last modified: | 2021-10-27 | | Method: | X-RAY DIFFRACTION (1.75 Å) | | Cite: | The catalytic domain of Escherichia coli Lon protease has a unique fold and a Ser-Lys dyad in the active site
J.Biol.Chem., 279, 2004
|
|
5VLB
 
 | |
5VET
 
 | | PHOSPHOLIPASE A2, RE-REFINEMENT OF THE PDB STRUCTURE 1JQ8 WITHOUT THE PUTATIVE COMPLEXED OLIGOPEPTIDE | | Descriptor: | Phospholipase A2 VRV-PL-VIIIa | | Authors: | Wlodawer, A, Dauter, Z, Minor, W, Stanfield, R, Porebski, P, Jaskolski, M, Pozharski, E, Weichenberger, C.X, Rupp, B. | | Deposit date: | 2017-04-05 | | Release date: | 2018-06-20 | | Last modified: | 2022-04-13 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Detect, correct, retract: How to manage incorrect structural models.
FEBS J., 285, 2018
|
|
5VEP
 
 | | MOUSE KYNURENINE AMINOTRANSFERASE III, RE-REFINEMENT OF THE PDB STRUCTURE 3E2F | | Descriptor: | 4-(2-HYDROXYETHYL)-1-PIPERAZINE ETHANESULFONIC ACID, CALCIUM ION, DI(HYDROXYETHYL)ETHER, ... | | Authors: | Wlodawer, A, Dauter, Z, Minor, W, Stanfield, R, Porebski, P, Jaskolski, M, Pozharski, E, Weichenberger, C.X, Rupp, B. | | Deposit date: | 2017-04-05 | | Release date: | 2017-11-29 | | Last modified: | 2022-04-13 | | Method: | X-RAY DIFFRACTION (2.59 Å) | | Cite: | Detect, correct, retract: How to manage incorrect structural models.
FEBS J., 285, 2018
|
|
5VF5
 
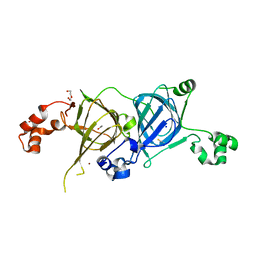 | | Crystal structure of the vicilin from Solanum melongena, re-refinement | | Descriptor: | ACETATE ION, COPPER (II) ION, DI(HYDROXYETHYL)ETHER, ... | | Authors: | Porebski, P.J, Wlodawer, A, Dauter, Z, Minor, W, Stanfield, R, Jaskolski, M, Pozharski, E, Weichenberger, C.X, Rupp, B. | | Deposit date: | 2017-04-06 | | Release date: | 2017-12-06 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (1.49 Å) | | Cite: | Detect, correct, retract: How to manage incorrect structural models.
FEBS J., 285, 2018
|
|
