5XF9
 
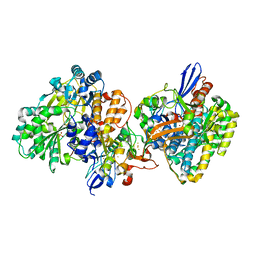 | | Crystal structure of NAD+-reducing [NiFe]-hydrogenase in the air-oxidized state | | Descriptor: | CARBONMONOXIDE-(DICYANO) IRON, FE2/S2 (INORGANIC) CLUSTER, FLAVIN MONONUCLEOTIDE, ... | | Authors: | Shomura, Y, Taketa, M, Nakashima, H, Tai, H, Nakagawa, H, Ikeda, Y, Ishii, M, Igarashi, Y, Nishihara, H, Yoon, K.S, Ogo, S, Hirota, S, Higuchi, Y. | | Deposit date: | 2017-04-09 | | Release date: | 2017-08-23 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (2.58 Å) | | Cite: | Structural basis of the redox switches in the NAD(+)-reducing soluble [NiFe]-hydrogenase
Science, 357, 2017
|
|
5XLF
 
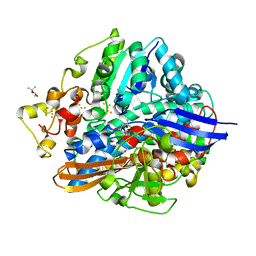 | | Crystal structure of aerobically purified and aerobically crystallized D. vulgaris Miyazaki F [NiFe]-hydrogenase | | Descriptor: | (4S)-2-METHYL-2,4-PENTANEDIOL, FE3-S4 CLUSTER, IRON/SULFUR CLUSTER, ... | | Authors: | Nishikawa, K, Mochida, S, Hiromoto, T, Shibata, N, Higuchi, Y. | | Deposit date: | 2017-05-10 | | Release date: | 2018-06-06 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (1.71 Å) | | Cite: | Ni-elimination from the active site of the standard [NiFe]‐hydrogenase upon oxidation by O2.
J. Inorg. Biochem., 177, 2017
|
|
5XED
 
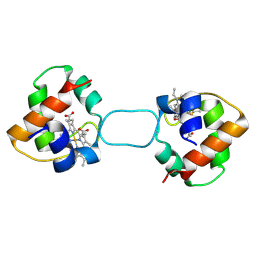 | | Heterodimer constructed from M61A PA cyt c551-HT cyt c552 and HT cyt c552-PA cyt c551 chimeric proteins | | Descriptor: | Cytochrome c-551,Cytochrome c-552, Cytochrome c-552,Cytochrome c-551, HEME C | | Authors: | Zhang, M, Nakanishi, T, Yamanaka, M, Nagao, S, Yanagisawa, S, Shomura, Y, Shibata, N, Ogura, T, Higuchi, Y, Hirota, S. | | Deposit date: | 2017-04-04 | | Release date: | 2017-08-09 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (1.55 Å) | | Cite: | Rational Design of Domain-Swapping-Based c-Type Cytochrome Heterodimers by Using Chimeric Proteins.
Chembiochem, 18, 2017
|
|
3AEY
 
 | |
5AUR
 
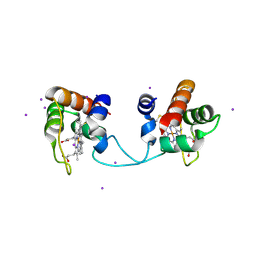 | | Hydrogenobacter thermophilus cytochrome c552 dimer formed by domain swapping at N-terminal region | | Descriptor: | Cytochrome c-552, HEME C, IODIDE ION | | Authors: | Ren, C, Nagao, S, Yamanaka, M, Kamikubo, H, Komori, H, Shomura, Y, Higuchi, Y, Hirota, S. | | Deposit date: | 2015-06-08 | | Release date: | 2015-10-21 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (1.26 Å) | | Cite: | Oligomerization enhancement and two domain swapping mode detection for thermostable cytochrome c552via the elongation of the major hinge loop.
Mol Biosyst, 11, 2015
|
|
3AEX
 
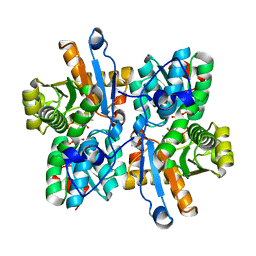 | |
5AUS
 
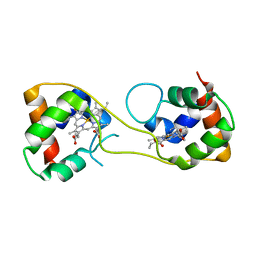 | | Hydrogenobacter thermophilus cytochrome c552 dimer formed by domain swapping at C-terminal region | | Descriptor: | Cytochrome c-552, HEME C | | Authors: | Ren, C, Nagao, S, Yamanaka, M, Komori, H, Shomura, Y, Higuchi, Y, Hirota, S. | | Deposit date: | 2015-06-08 | | Release date: | 2015-10-21 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (1.3 Å) | | Cite: | Oligomerization enhancement and two domain swapping mode detection for thermostable cytochrome c552via the elongation of the major hinge loop.
Mol Biosyst, 11, 2015
|
|
3ALB
 
 | | Cyclic Lys48-linked tetraubiquitin | | Descriptor: | SULFATE ION, ubiquitin | | Authors: | Satoh, T, Sakata, E, Yamamoto, S, Yamaguchi, Y, Sumiyoshi, A, Wakatsuki, S, Kato, K. | | Deposit date: | 2010-07-29 | | Release date: | 2010-08-25 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | Crystal structure of cyclic Lys48-linked tetraubiquitin
Biochem.Biophys.Res.Commun., 2010
|
|
3AN1
 
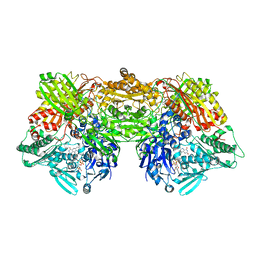 | | Crystal structure of rat D428A mutant, urate bound form | | Descriptor: | BICARBONATE ION, CALCIUM ION, FE2/S2 (INORGANIC) CLUSTER, ... | | Authors: | Okamoto, K, Kawaguchi, Y, Eger, B.T, Pai, E.F, Nishino, T. | | Deposit date: | 2010-08-27 | | Release date: | 2010-12-01 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (1.73 Å) | | Cite: | Crystal Structures of Urate Bound Form of Xanthine Oxidoreductase: Substrate Orientation and Structure of the Key Reaction Intermediate
J.Am.Chem.Soc., 132, 2010
|
|
5XVC
 
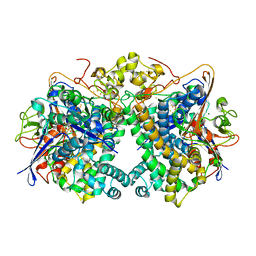 | | [NiFe]-hydrogenase (Hyb-type) from Citrobacter sp. S-77 in a ferricyanide-oxidized condition | | Descriptor: | DI(HYDROXYETHYL)ETHER, FE3-S4 CLUSTER, FE4-S4-O CLUSTER, ... | | Authors: | Nishikawa, K, Matsuura, H, Muhd Noor, N.D, Tai, H, Hirota, S, Kim, J, Kang, J, Tateno, M, Yoon, K.S, Ogo, S, Shomura, Y, Higuchi, Y. | | Deposit date: | 2017-06-27 | | Release date: | 2018-06-27 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (2.05 Å) | | Cite: | Redox-dependent conformational changes of a proximal [4Fe-4S] cluster in Hyb-type [NiFe]-hydrogenase to protect the active site from O2.
Chem.Commun.(Camb.), 54, 2018
|
|
3AT9
 
 | |
3AQP
 
 | | Crystal structure of SecDF, a translocon-associated membrane protein, from Thermus thrmophilus | | Descriptor: | Probable SecDF protein-export membrane protein | | Authors: | Tsukazaki, T, Mori, H, Echizen, Y, Ishitani, R, Fukai, S, Tanaka, T, Perederina, A, Vassylyev, D.G, Kohno, T, Ito, K, Nureki, O. | | Deposit date: | 2010-11-16 | | Release date: | 2011-05-18 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (3.3 Å) | | Cite: | Structure and function of a membrane component SecDF that enhances protein export
Nature, 474, 2011
|
|
3ATB
 
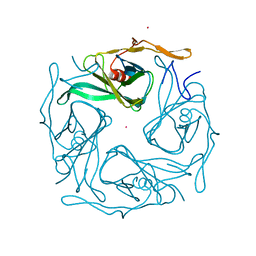 | |
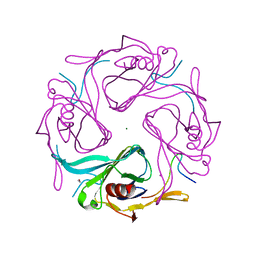
3AUW
 
 | |
3ATF
 
 | |
5XEC
 
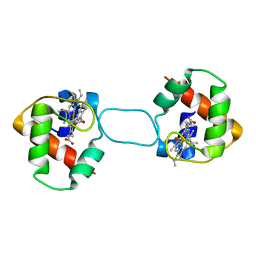 | | Heterodimer constructed from PA cyt c551-HT cyt c552 and HT cyt c552-PA cyt c551 chimeric proteins | | Descriptor: | Cytochrome c-551,Cytochrome c-552, Cytochrome c-552,Cytochrome c-551, HEME C | | Authors: | Zhang, M, Nakanishi, T, Yamanaka, M, Nagao, S, Yanagisawa, S, Shomura, Y, Shibata, N, Ogura, T, Higuchi, Y, Hirota, S. | | Deposit date: | 2017-04-04 | | Release date: | 2017-08-09 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (1.1 Å) | | Cite: | Rational Design of Domain-Swapping-Based c-Type Cytochrome Heterodimers by Using Chimeric Proteins.
Chembiochem, 18, 2017
|
|
3AT8
 
 | |
3ATD
 
 | |
6EXP
 
 | | Crystal structure of the SIRV3 AcrID1 (gp02) anti-CRISPR protein | | Descriptor: | SIRV3 AcrID1 (gp02) anti-CRISPR protein | | Authors: | He, F, Bhoobalan-Chitty, Y, Van, L.B, Kjeldsen, A.L, Dedola, M, Makarova, K.S, Koonin, E.V, Brodersen, D.E, Peng, X. | | Deposit date: | 2017-11-08 | | Release date: | 2018-01-31 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (1.93 Å) | | Cite: | Anti-CRISPR proteins encoded by archaeal lytic viruses inhibit subtype I-D immunity.
Nat Microbiol, 3, 2018
|
|
1WYW
 
 | | Crystal Structure of SUMO1-conjugated thymine DNA glycosylase | | Descriptor: | CHLORIDE ION, G/T mismatch-specific thymine DNA glycosylase, MAGNESIUM ION, ... | | Authors: | Baba, D, Maita, N, Jee, J.G, Uchimura, Y, Saitoh, H, Sugasawa, K, Hanaoka, F, Tochio, H, Hiroaki, H, Shirakawa, M. | | Deposit date: | 2005-02-17 | | Release date: | 2005-06-21 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Crystal structure of thymine DNA glycosylase conjugated to SUMO-1.
Nature, 435, 2005
|
|
5B5V
 
 | | Structure of full-length MOB1b | | Descriptor: | CHLORIDE ION, MOB kinase activator 1B, ZINC ION | | Authors: | KIM, S.-Y, Tachioka, Y, Mori, T, Hakoshima, T. | | Deposit date: | 2016-05-24 | | Release date: | 2016-07-06 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (2.193 Å) | | Cite: | Structural basis for autoinhibition and its relief of MOB1 in the Hippo pathway
Sci Rep, 6, 2016
|
|
5B7E
 
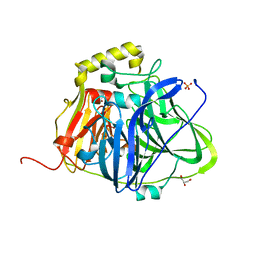 | | Structure of perdeuterated CueO | | Descriptor: | Blue copper oxidase CueO, COPPER (II) ION, CU-O-CU LINKAGE, ... | | Authors: | Akter, M, Higuchi, Y, Shibata, N. | | Deposit date: | 2016-06-07 | | Release date: | 2016-10-19 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (1.42 Å) | | Cite: | Biochemical, spectroscopic and X-ray structural analysis of deuterated multicopper oxidase CueO prepared from a new expression construct for neutron crystallography
Acta Crystallogr.,Sect.F, 72, 2016
|
|
5B1X
 
 | |
5AZX
 
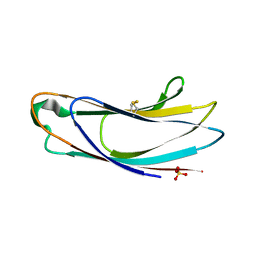 | | Crystal structure of p24delta1 GOLD domain (native 1) | | Descriptor: | SULFATE ION, Transmembrane emp24 domain-containing protein 10 | | Authors: | Nagae, M, Yamaguchi, Y. | | Deposit date: | 2015-10-23 | | Release date: | 2016-09-14 | | Last modified: | 2020-02-26 | | Method: | X-RAY DIFFRACTION (1.58 Å) | | Cite: | 3D Structure and Interaction of p24 beta and p24 delta Golgi Dynamics Domains: Implication for p24 Complex Formation and Cargo Transport
J.Mol.Biol., 428, 2016
|
|
5B5W
 
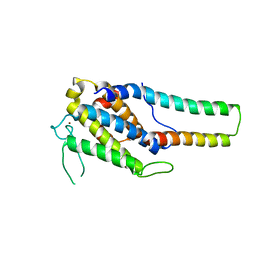 | | Crystal structure of MOB1-LATS1 NTR domain complex | | Descriptor: | MOB kinase activator 1B, Serine/threonine-protein kinase LATS1, ZINC ION | | Authors: | KIM, S.-Y, Tachioka, Y, Mori, T, Hakoshima, T. | | Deposit date: | 2016-05-24 | | Release date: | 2016-07-06 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.957 Å) | | Cite: | Structural basis for autoinhibition and its relief of MOB1 in the Hippo pathway
Sci Rep, 6, 2016
|
|
