5YYP
 
 | | Structure K137A thaumatin | | Descriptor: | GLYCEROL, L(+)-TARTARIC ACID, Preprothaumatin I | | Authors: | Masuda, T, Kigo, S, Mitsumoto, M, Ohta, K, Suzuki, M, Mikami, B, Kitabatake, N, Tani, F. | | Deposit date: | 2017-12-10 | | Release date: | 2018-03-21 | | Last modified: | 2024-11-13 | | Method: | X-RAY DIFFRACTION (1.01 Å) | | Cite: | Positive Charges on the Surface of Thaumatin Are Crucial for the Multi-Point Interaction with the Sweet Receptor.
Front Mol Biosci, 5, 2018
|
|
5YYQ
 
 | | Structure K78A thaumatin | | Descriptor: | GLYCEROL, L(+)-TARTARIC ACID, Preprothaumatin I | | Authors: | Masuda, T, Kigo, S, Mitsumoto, M, Ohta, K, Suzuki, M, Mikami, B, Kitabatake, N, Tani, F. | | Deposit date: | 2017-12-10 | | Release date: | 2018-03-21 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (1.07 Å) | | Cite: | Positive Charges on the Surface of Thaumatin Are Crucial for the Multi-Point Interaction with the Sweet Receptor.
Front Mol Biosci, 5, 2018
|
|
1WR6
 
 | | Crystal structure of GGA3 GAT domain in complex with ubiquitin | | Descriptor: | ADP-ribosylation factor binding protein GGA3, ubiquitin | | Authors: | Kawasaki, M, Shiba, T, Shiba, Y, Yamaguchi, Y, Matsugaki, N, Igarashi, N, Suzuki, M, Kato, R, Kato, K, Nakayama, K, Wakatsuki, S. | | Deposit date: | 2004-10-12 | | Release date: | 2005-06-28 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Molecular mechanism of ubiquitin recognition by GGA3 GAT domain.
Genes Cells, 10, 2005
|
|
5B35
 
 | | Serial Femtosecond Crystallography (SFX) of Ground State Bacteriorhodopsin Crystallized from Bicelles Determined Using 7-keV X-ray Free Electron Laser (XFEL) at SACLA | | Descriptor: | (3R,5S,7R,8R,9S,10S,12S,13R,14S,17R)-10,13-dimethyl-17-[(2R)-pentan-2-yl]-2,3,4,5,6,7,8,9,11,12,14,15,16,17-tetradecahydro-1H-cyclopenta[a]phenanthrene-3,7,12-triol, Bacteriorhodopsin, DECANE, ... | | Authors: | Mizohata, E, Nakane, T, Suzuki, M. | | Deposit date: | 2016-02-10 | | Release date: | 2016-11-09 | | Last modified: | 2024-11-20 | | Method: | X-RAY DIFFRACTION (2.35 Å) | | Cite: | Membrane protein structure determination by SAD, SIR, or SIRAS phasing in serial femtosecond crystallography using an iododetergent
Proc.Natl.Acad.Sci.USA, 113, 2016
|
|
6AD9
 
 | | Crystal Structure of PPARgamma Ligand Binding Domain in complex with dibenzooxepine derivative compound-9 | | Descriptor: | 12-mer peptide from Peroxisome proliferator-activated receptor gamma coactivator 1-alpha, 3-[(1E)-1-{8-[(4-methyl-2-propyl-1H-benzimidazol-1-yl)methyl]dibenzo[b,e]oxepin-11(6H)-ylidene}ethyl]-1,2,4-oxadiazol-5(4H)-one, Peroxisome proliferator-activated receptor gamma | | Authors: | Takahashi, Y, Suzuki, M, Yamamoto, K, Saito, J. | | Deposit date: | 2018-07-31 | | Release date: | 2018-11-14 | | Last modified: | 2024-03-27 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Development of Dihydrodibenzooxepine Peroxisome Proliferator-Activated Receptor (PPAR) Gamma Ligands of a Novel Binding Mode as Anticancer Agents: Effective Mimicry of Chiral Structures by Olefinic E/ Z-Isomers.
J. Med. Chem., 61, 2018
|
|
5YYR
 
 | | Structure K106A thaumatin | | Descriptor: | GLYCEROL, L(+)-TARTARIC ACID, Preprothaumatin I | | Authors: | Masuda, T, Kigo, S, Ohta, K, Mitsumoto, M, Mikami, B, Suzuki, M, Kitabatake, N, Tani, F. | | Deposit date: | 2017-12-10 | | Release date: | 2018-03-21 | | Last modified: | 2024-11-13 | | Method: | X-RAY DIFFRACTION (1.07 Å) | | Cite: | Positive Charges on the Surface of Thaumatin Are Crucial for the Multi-Point Interaction with the Sweet Receptor.
Front Mol Biosci, 5, 2018
|
|
5D4I
 
 | | Intact nitrite complex of a copper nitrite reductase determined by serial femtosecond crystallography | | Descriptor: | COPPER (II) ION, Copper-containing nitrite reductase, NITRITE ION | | Authors: | Fukuda, Y, Tse, K.M, Nakane, T, Nakatsu, T, Suzuki, M, Sugahara, M, Inoue, S, Masuda, T, Yumoto, F, Matsugaki, N, Nango, E, Tono, K, Joti, Y, Kameshima, T, Song, C, Hatsui, T, Yabashi, M, Nureki, O, Murphy, M.E.P, Inoue, T, Iwata, S, Mizohata, E. | | Deposit date: | 2015-08-07 | | Release date: | 2016-03-09 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Redox-coupled proton transfer mechanism in nitrite reductase revealed by femtosecond crystallography
Proc.Natl.Acad.Sci.USA, 113, 2016
|
|
2D05
 
 | | Chitosanase From Bacillus circulans mutant K218P | | Descriptor: | Chitosanase, SULFATE ION | | Authors: | Fukamizo, T, Amano, S, Yamaguchi, K, Yoshikawa, T, Katsumi, T, Saito, J, Suzuki, M, Miki, K, Nagata, Y, Ando, A. | | Deposit date: | 2005-07-25 | | Release date: | 2005-12-06 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Bacillus circulans MH-K1 Chitosanase: Amino Acid Residues Responsible for Substrate Binding
J.Biochem.(Tokyo), 138, 2005
|
|
5B1D
 
 | |
5B1F
 
 | |
5B1E
 
 | |
5B1G
 
 | |
1HI3
 
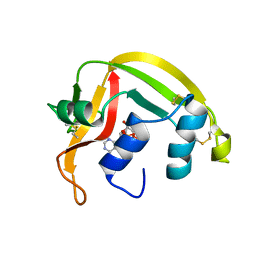 | | Eosinophil-derived Neurotoxin (EDN) - Adenosine 2'-5'-Diphosphate Complex | | Descriptor: | ADENOSINE-2'-5'-DIPHOSPHATE, EOSINOPHIL-DERIVED NEUROTOXIN | | Authors: | Leonidas, D.D, Boix, E, Prill, R, Suzuki, M, Turton, R, Minson, K, Swaminathan, G.J, Youle, R.J, Acharya, K.R. | | Deposit date: | 2001-01-02 | | Release date: | 2001-05-31 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Mapping the Ribonucleolytic Active Site of Eosinophil-Derived Neurotoxin (Edn): High Resolution Crystal Structures of Edn Complexes with Adenylic Nucleotide Inhibitors
J.Biol.Chem., 276, 2001
|
|
7W05
 
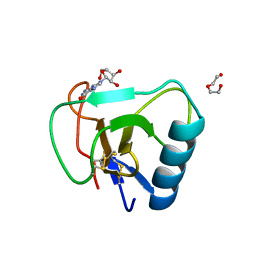 | | 12 mutant Ribonuclease from Hericium erinaceus GMP binding form | | Descriptor: | DI(HYDROXYETHYL)ETHER, GUANOSINE, Ribonuclease T1 | | Authors: | Takebe, K, Chida, T, Suzuki, M, Itagaki, T, Morita, Y, Uzawa, N, Kobayashi, H. | | Deposit date: | 2021-11-17 | | Release date: | 2022-11-23 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (1.29 Å) | | Cite: | 12 mutant Ribonuclease from Hericium erinaceus GMP binding form
To Be Published
|
|
1HI2
 
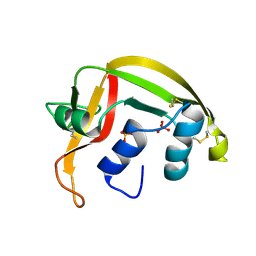 | | Eosinophil-derived Neurotoxin (EDN) - Sulphate Complex | | Descriptor: | EOSINOPHIL-DERIVED NEUROTOXIN, SULFATE ION | | Authors: | Leonidas, D.D, Boix, E, Prill, R, Suzuki, M, Turton, R, Minson, K, Swaminathan, G.J, Youle, R.J, Acharya, K.R. | | Deposit date: | 2001-01-02 | | Release date: | 2001-05-31 | | Last modified: | 2024-11-20 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Mapping the Ribonucleolytic Active Site of Eosinophil-Derived Neurotoxin (Edn): High Resolution Crystal Structures of Edn Complexes with Adenylic Nucleotide Inhibitors
J.Biol.Chem., 276, 2001
|
|
1HI5
 
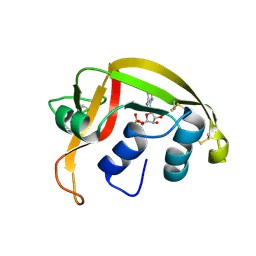 | | Eosinophil-derived Neurotoxin (EDN) - Adenosine-5'-Diphosphate Complex | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, EOSINOPHIL-DERIVED NEUROTOXIN | | Authors: | Leonidas, D.D, Boix, E, Prill, R, Suzuki, M, Turton, R, Minson, K, Swaminathan, G.J, Youle, R.J, Acharya, K.R. | | Deposit date: | 2001-01-02 | | Release date: | 2001-05-31 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Mapping the Ribonucleolytic Active Site of Eosinophil-Derived Neurotoxin (Edn): High Resolution Crystal Structures of Edn Complexes with Adenylic Nucleotide Inhibitors
J.Biol.Chem., 276, 2001
|
|
1HI4
 
 | | Eosinophil-derived Neurotoxin (EDN) - Adenosien-3'-5'-Diphosphate Complex | | Descriptor: | ADENOSINE-3'-5'-DIPHOSPHATE, EOSINOPHIL-DERIVED NEUROTOXIN | | Authors: | Leonidas, D.D, Boix, E, Prill, R, Suzuki, M, Turton, R, Minson, K, Swaminathan, G.J, Youle, R.J, Acharya, K.R. | | Deposit date: | 2001-01-02 | | Release date: | 2001-05-31 | | Last modified: | 2024-11-13 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Mapping the Ribonucleolytic Active Site of Eosinophil-Derived Neurotoxin (Edn): High Resolution Crystal Structures of Edn Complexes with Adenylic Nucleotide Inhibitors
J.Biol.Chem., 276, 2001
|
|
7XJB
 
 | | Rat-COMT, opicapone,SAM and Mg bond | | Descriptor: | CHLORIDE ION, Catechol O-methyltransferase, MAGNESIUM ION, ... | | Authors: | Takebe, K, Iijima, H, Suzuki, M, Kuwada-Kusunose, T. | | Deposit date: | 2022-04-15 | | Release date: | 2023-05-31 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Structural and Computational Analyses of the Unique Interactions of Opicapone in the Binding Pocket of Catechol O -Methyltransferase: A Crystallographic Study and Fragment Molecular Orbital Analyses.
J.Chem.Inf.Model., 63, 2023
|
|
7XGI
 
 | | COMT SAH Mg opicapone complex | | Descriptor: | Catechol O-methyltransferase, MAGNESIUM ION, Opicapone, ... | | Authors: | Takebe, K, Kuwada-Kusunose, T, Suzuki, M, Iijima, H. | | Deposit date: | 2022-04-04 | | Release date: | 2023-04-12 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Structural and Computational Analyses of the Unique Interactions of Opicapone in the Binding Pocket of Catechol O -Methyltransferase: A Crystallographic Study and Fragment Molecular Orbital Analyses.
J.Chem.Inf.Model., 63, 2023
|
|
2GCC
 
 | | SOLUTION STRUCTURE OF THE GCC-BOX BINDING DOMAIN, NMR, MINIMIZED MEAN STRUCTURE | | Descriptor: | ATERF1 | | Authors: | Allen, M.D, Yamasaki, K, Ohme-Takagi, M, Tateno, M, Suzuki, M. | | Deposit date: | 1998-03-13 | | Release date: | 1999-03-23 | | Last modified: | 2024-05-29 | | Method: | SOLUTION NMR | | Cite: | A novel mode of DNA recognition by a beta-sheet revealed by the solution structure of the GCC-box binding domain in complex with DNA.
EMBO J., 17, 1998
|
|
1ISP
 
 | | Crystal structure of Bacillus subtilis lipase at 1.3A resolution | | Descriptor: | GLYCEROL, lipase | | Authors: | Kawasaki, K, Kondo, H, Suzuki, M, Ohgiya, S, Tsuda, S. | | Deposit date: | 2001-12-19 | | Release date: | 2002-12-19 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (1.3 Å) | | Cite: | Alternate conformations observed in catalytic serine of Bacillus subtilis lipase determined at 1.3 A resolution.
Acta Crystallogr.,Sect.D, 58, 2002
|
|
3WHO
 
 | | X-ray-Crystallographic Structure of an RNase Po1 Exhibiting Anti-tumor Activity | | Descriptor: | Guanyl-specific ribonuclease Po1 | | Authors: | Kobayashi, H, Katsurtani, T, Hara, Y, Motoyoshi, N, Itagaki, T, Akita, F, Higashiura, A, Yamada, Y, Suzuki, M, Inokuchi, N. | | Deposit date: | 2013-08-30 | | Release date: | 2014-07-02 | | Last modified: | 2024-11-13 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | X-ray crystallographic structure of RNase Po1 that exhibits anti-tumor activity.
Biol.Pharm.Bull., 37, 2014
|
|
1WZV
 
 | | Crystal Structure of UbcH8 | | Descriptor: | Ubiquitin-conjugating enzyme E2 L6 | | Authors: | Mizushima, T, Suzuki, M, Teshima, N, Yamane, T, Murata, S, Tanaka, K. | | Deposit date: | 2005-03-10 | | Release date: | 2005-03-22 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Crystal Structure of UbcH8
To be Published
|
|
1WZW
 
 | | Crystal Structure of UbcH8 | | Descriptor: | Ubiquitin-conjugating enzyme E2 L6 | | Authors: | Mizushima, T, Suzuki, M, Teshima, N, Yamane, T, Murata, S, Tanaka, K. | | Deposit date: | 2005-03-10 | | Release date: | 2005-03-22 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Crystal Structure of UbcH8
To be Published
|
|
2E1A
 
 | | crystal structure of FFRP-DM1 | | Descriptor: | 75aa long hypothetical regulatory protein AsnC, SELENOMETHIONINE | | Authors: | Koike, H, Suzuki, M. | | Deposit date: | 2006-10-19 | | Release date: | 2007-09-18 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | A Structural Code for Discriminating between Transcription Signals Revealed by the Feast/Famine Regulatory Protein DM1 in Complex with Ligands
Structure, 15, 2007
|
|
