3R1Q
 
 | | CDK2 in complex with inhibitor KVR-1-102 | | Descriptor: | 1,2-ETHANEDIOL, 4-chloro-2-{[(6-chloropyridin-3-yl)methyl]amino}-5-nitrobenzamide, Cyclin-dependent kinase 2 | | Authors: | Betzi, S, Alam, R, Han, H, Becker, A, Schonbrunn, E. | | Deposit date: | 2011-03-11 | | Release date: | 2012-08-08 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | Structure-guided optimization of novel CDK2 inhibitors discovered by high-throughput screening
To be Published
|
|
3R73
 
 | | CDK2 in complex with inhibitor KVR-1-164 | | Descriptor: | 1,2-ETHANEDIOL, 4-[(3-aminopropyl)amino]-5-nitro-2-[(pyridin-3-ylmethyl)amino]benzamide, Cyclin-dependent kinase 2 | | Authors: | Betzi, S, Alam, R, Han, H, Becker, A, Schonbrunn, E. | | Deposit date: | 2011-03-22 | | Release date: | 2012-08-08 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Structure-guided optimization of novel CDK2 inhibitors discovered by high-throughput screening
To be Published
|
|
3R28
 
 | | CDK2 in complex with inhibitor KVR-1-140 | | Descriptor: | 1,2-ETHANEDIOL, 4-chloro-5-nitro-2-[(3,4,5-trifluorobenzyl)amino]benzamide, Cyclin-dependent kinase 2 | | Authors: | Betzi, S, Alam, R, Han, H, Becker, A, Schonbrunn, E. | | Deposit date: | 2011-03-14 | | Release date: | 2012-08-08 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (1.75 Å) | | Cite: | Structure-guided optimization of novel CDK2 inhibitors discovered by high-throughput screening
To be Published
|
|
3R7I
 
 | | CDK2 in complex with inhibitor KVR-1-74 | | Descriptor: | 1,2-ETHANEDIOL, 4-{[(2-carbamoyl-4-nitrophenyl)amino]methyl}benzoic acid, Cyclin-dependent kinase 2 | | Authors: | Betzi, S, Alam, R, Han, H, Becker, A, Schonbrunn, E. | | Deposit date: | 2011-03-22 | | Release date: | 2012-08-08 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | Structure-guided optimization of novel CDK2 inhibitors discovered by high-throughput screening
To be Published
|
|
3UNJ
 
 | | CDK2 in complex with inhibitor YL1-038-31 | | Descriptor: | 4-{[4-(phenylamino)pyrimidin-2-yl]amino}benzoic acid, Cyclin-dependent kinase 2, PHOSPHATE ION | | Authors: | Zhu, J.-Y, Martin, M.P, Alam, R, Schonbrunn, E. | | Deposit date: | 2011-11-15 | | Release date: | 2012-01-25 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (1.9001 Å) | | Cite: | A Novel Mechanism by Which Small Molecule Inhibitors Induce the DFG Flip in Aurora A.
Acs Chem.Biol., 7, 2012
|
|
3RM6
 
 | | CDK2 in complex with inhibitor KVR-2-80 | | Descriptor: | 1,2-ETHANEDIOL, 2-{[(2-aminopyrimidin-5-yl)methyl]amino}-5-nitro-4-{[2-(piperazin-1-yl)ethyl]amino}benzamide, Cyclin-dependent kinase 2 | | Authors: | Betzi, S, Alam, R, Han, H, Becker, A, Schonbrunn, E. | | Deposit date: | 2011-04-20 | | Release date: | 2012-08-08 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Structure-guided optimization of novel CDK2 inhibitors discovered by high-throughput screening
To be Published
|
|
3RAI
 
 | | CDK2 in complex with inhibitor KVR-1-160 | | Descriptor: | 1,2-ETHANEDIOL, 4-{[3-(morpholin-4-yl)propyl]amino}-5-nitro-2-[(pyridin-3-ylmethyl)amino]benzamide, Cyclin-dependent kinase 2 | | Authors: | Betzi, S, Alam, R, Han, H, Becker, A, Schonbrunn, E. | | Deposit date: | 2011-03-28 | | Release date: | 2012-08-08 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Structure-guided optimization of novel CDK2 inhibitors discovered by high-throughput screening
To be Published
|
|
3R1Y
 
 | | CDK2 in complex with inhibitor KVR-1-134 | | Descriptor: | 1,2-ETHANEDIOL, 2-[(pyridin-3-ylmethyl)amino]benzamide, Cyclin-dependent kinase 2 | | Authors: | Betzi, S, Alam, R, Han, H, Becker, A, Schonbrunn, E. | | Deposit date: | 2011-03-11 | | Release date: | 2012-08-08 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Structure-guided optimization of novel CDK2 inhibitors discovered by high-throughput screening
To be Published
|
|
3R7Y
 
 | | CDK2 in complex with inhibitor KVR-2-88 | | Descriptor: | 1,2-ETHANEDIOL, 2-{[(2-aminopyrimidin-5-yl)methyl]amino}-4-(morpholin-4-yl)-5-nitrobenzamide, Cyclin-dependent kinase 2 | | Authors: | Betzi, S, Alam, R, Han, H, Becker, A, Schonbrunn, E. | | Deposit date: | 2011-03-23 | | Release date: | 2012-08-08 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Structure-guided optimization of novel CDK2 inhibitors discovered by high-throughput screening
To be Published
|
|
3R8L
 
 | | CDK2 in complex with inhibitor L3-4 | | Descriptor: | (5R)-5-tert-butyl-4,5,6,7-tetrahydro-1H-indazole-3-carbohydrazide, Cyclin-dependent kinase 2 | | Authors: | Betzi, S, Alam, R, Han, H, Becker, A, Schonbrunn, E. | | Deposit date: | 2011-03-24 | | Release date: | 2012-08-08 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Structure-guided optimization of novel CDK2 inhibitors discovered by high-throughput screening
To be Published
|
|
3ROY
 
 | | CDK2 in complex with inhibitor KVR-1-154 | | Descriptor: | 1,2-ETHANEDIOL, 4-(hexylamino)-5-nitro-2-[(pyridin-3-ylmethyl)amino]benzamide, Cyclin-dependent kinase 2 | | Authors: | Betzi, S, Alam, R, Han, H, Becker, A, Schonbrunn, E. | | Deposit date: | 2011-04-26 | | Release date: | 2012-08-08 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (1.75 Å) | | Cite: | Structure-guided optimization of novel CDK2 inhibitors discovered by high-throughput screening
To be Published
|
|
3R8P
 
 | | CDK2 in complex with inhibitor NSK-MC1-6 | | Descriptor: | (5R)-5-propyl-4,5,6,7-tetrahydro-1H-indazole-3-carbohydrazide, Cyclin-dependent kinase 2 | | Authors: | Betzi, S, Alam, R, Han, H, Becker, A, Schonbrunn, E. | | Deposit date: | 2011-03-24 | | Release date: | 2012-08-08 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Structure-guided optimization of novel CDK2 inhibitors discovered by high-throughput screening
To be Published
|
|
3R7E
 
 | | CDK2 in complex with inhibitor KVR-1-67 | | Descriptor: | 1,2-ETHANEDIOL, 5-nitro-2-[(pyrazin-2-ylmethyl)amino]benzamide, Cyclin-dependent kinase 2 | | Authors: | Betzi, S, Alam, R, Han, H, Becker, A, Schonbrunn, E. | | Deposit date: | 2011-03-22 | | Release date: | 2012-08-08 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Structure-guided optimization of novel CDK2 inhibitors discovered by high-throughput screening
To be Published
|
|
3R8M
 
 | | CDK2 in complex with inhibitor L3-3 | | Descriptor: | 1H-indazole-3-carbohydrazide, Cyclin-dependent kinase 2, PHOSPHATE ION | | Authors: | Betzi, S, Alam, R, Han, H, Becker, A, Schonbrunn, E. | | Deposit date: | 2011-03-24 | | Release date: | 2012-08-08 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Structure-guided optimization of novel CDK2 inhibitors discovered by high-throughput screening
To be Published
|
|
4JO5
 
 | | CBM3a-L domain with flanking linkers from scaffoldin cipA of cellulosome of Clostridium thermocellum | | Descriptor: | 1-METHYLIMIDAZOLE, CALCIUM ION, CHLORIDE ION, ... | | Authors: | Shimon, L.J.W, Frolow, F, Bayer, E.A, Yaniv, O, Lamed, R, Morag, E. | | Deposit date: | 2013-03-16 | | Release date: | 2013-07-17 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (1.98 Å) | | Cite: | Structure of a family 3a carbohydrate-binding module from the cellulosomal scaffoldin CipA of Clostridium thermocellum with flanking linkers: implications for cellulosome structure.
Acta Crystallogr.,Sect.F, 69, 2013
|
|
1NBC
 
 | | BACTERIAL TYPE 3A CELLULOSE-BINDING DOMAIN | | Descriptor: | CALCIUM ION, CELLULOSOMAL SCAFFOLDING PROTEIN A | | Authors: | Tormo, J, Lamed, R, Steitz, T.A. | | Deposit date: | 1996-09-10 | | Release date: | 1997-09-26 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (1.75 Å) | | Cite: | Crystal structure of a bacterial family-III cellulose-binding domain: a general mechanism for attachment to cellulose.
EMBO J., 15, 1996
|
|
1PDA
 
 | | STRUCTURE OF PORPHOBILINOGEN DEAMINASE REVEALS A FLEXIBLE MULTIDOMAIN POLYMERASE WITH A SINGLE CATALYTIC SITE | | Descriptor: | 3-[5-{[3-(2-carboxyethyl)-4-(carboxymethyl)-5-methyl-1H-pyrrol-2-yl]methyl}-4-(carboxymethyl)-1H-pyrrol-3-yl]propanoic acid, ACETIC ACID, PORPHOBILINOGEN DEAMINASE | | Authors: | Louie, G.V, Brownlie, P.D, Lambert, R, Cooper, J.B, Blundell, T.L, Wood, S.P, Warren, M.J, Woodcock, S.C, Jordan, P.M. | | Deposit date: | 1992-11-17 | | Release date: | 1993-10-31 | | Last modified: | 2019-08-14 | | Method: | X-RAY DIFFRACTION (1.76 Å) | | Cite: | Structure of porphobilinogen deaminase reveals a flexible multidomain polymerase with a single catalytic site.
Nature, 359, 1992
|
|
1QZN
 
 | | Crystal Structure Analysis of a type II cohesin domain from the cellulosome of Acetivibrio cellulolyticus | | Descriptor: | cellulosomal scaffoldin adaptor protein B | | Authors: | Frolow, F, Noach, I, Rosenheck, S, Lamed, R, Qi, X, Shimon, L.J.W, Bayer, E.A. | | Deposit date: | 2003-09-17 | | Release date: | 2004-09-21 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Crystal structure of a type-II cohesin module from the Bacteroides cellulosolvens cellulosome reveals novel and distinctive secondary structural elements.
J.Mol.Biol., 348, 2005
|
|
2WO4
 
 | | 3b' carbohydrate-binding module from the Cel9V glycoside hydrolase from Clostridium thermocellum, in-house data | | Descriptor: | CALCIUM ION, CHLORIDE ION, GLYCOSIDE HYDROLASE, ... | | Authors: | Petkun, S, Jindou, S, Shimon, L.J.W, Bayer, E.A, Lamed, R, Frolow, F. | | Deposit date: | 2009-07-21 | | Release date: | 2009-12-29 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | Structure of a Family 3B' Carbohydrate-Binding Module from the Cel9V Glycoside Hydrolase from Clostridium Thermocellum: Structural Diversity and Implications for Carbohydrate Binding
Acta Crystallogr.,Sect.D, 66, 2010
|
|
2WNX
 
 | | 3b' carbohydrate-binding module from the Cel9V glycoside hydrolase from Clostridium thermocellum | | Descriptor: | CALCIUM ION, FORMIC ACID, GLYCOSIDE HYDROLASE, ... | | Authors: | Petkun, S, Jindou, S, Shimon, L.J.W, Bayer, E.A, Lamed, R, Frolow, F. | | Deposit date: | 2009-07-20 | | Release date: | 2009-12-29 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (1.31 Å) | | Cite: | Structure of a Family 3B' Carbohydrate-Binding Module from the Cel9V Glycoside Hydrolase from Clostridium Thermocellum: Structural Diversity and Implications for Carbohydrate Binding
Acta Crystallogr.,Sect.D, 66, 2010
|
|
2WOB
 
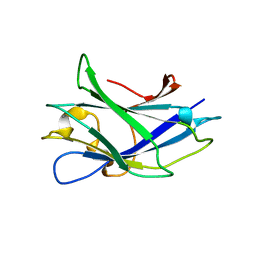 | | 3b' carbohydrate-binding module from the Cel9V glycoside hydrolase from Clostridium thermocellum. Orthorhombic structure | | Descriptor: | CALCIUM ION, GLYCOSIDE HYDROLASE, FAMILY 9 | | Authors: | Petkun, S, Jindou, S, Shimon, L.J.W, Bayer, E.A, Lamed, R, Frolow, F. | | Deposit date: | 2009-07-22 | | Release date: | 2009-12-29 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Structure of a Family 3B' Carbohydrate-Binding Module from the Cel9V Glycoside Hydrolase from Clostridium Thermocellum: Structural Diversity and Implications for Carbohydrate Binding
Acta Crystallogr.,Sect.D, 66, 2010
|
|
2XDH
 
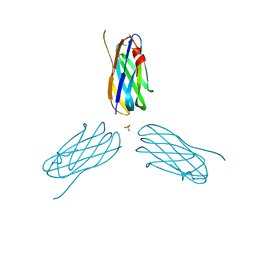 | | Non-cellulosomal cohesin from the hyperthermophilic archaeon Archaeoglobus fulgidus | | Descriptor: | CHLORIDE ION, COHESIN, MAGNESIUM ION, ... | | Authors: | Voronov-Goldman, M, Lamed, R, Noach, I, Borovok, I, Kwiat, M, Rosenheck, S, Shimon, L.J.W, Bayer, E.A, Frolow, F. | | Deposit date: | 2010-05-02 | | Release date: | 2010-05-26 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (1.96 Å) | | Cite: | Non-Cellulosomal Cohesin from the Hyperthermophilic Archaeon Archaeoglobus Fulgidus
Proteins, 79, 2011
|
|
4GCJ
 
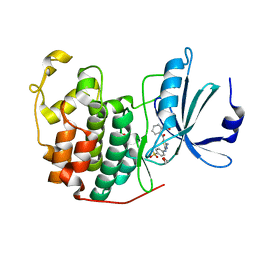 | | CDK2 in complex with inhibitor RC-3-89 | | Descriptor: | 1,2-ETHANEDIOL, 4-{[4-amino-5-(2-nitrobenzoyl)-1,3-thiazol-2-yl]amino}benzenesulfonamide, Cyclin-dependent kinase 2 | | Authors: | Betzi, S, Alam, R, Han, H, Becker, A, Schonbrunn, E. | | Deposit date: | 2012-07-30 | | Release date: | 2012-10-31 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (1.42 Å) | | Cite: | Development of highly potent and selective diaminothiazole inhibitors of cyclin-dependent kinases.
J.Med.Chem., 56, 2013
|
|
4QJY
 
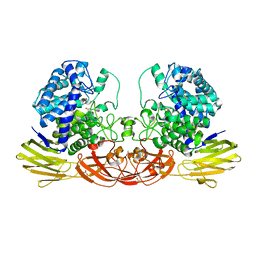 | | Crystal structure of native Ara127N, a GH127 beta-L-arabinofuranosidase from Geobacillus Stearothermophilus T6 | | Descriptor: | ACETATE ION, GH127 beta-L-arabinofuranosidase | | Authors: | Lansky, S, Salama, R, Dann, R, Shner, I, Manjasetty, B, Belrhali, H, Shoham, Y, Shoham, G. | | Deposit date: | 2014-06-05 | | Release date: | 2015-06-10 | | Method: | X-RAY DIFFRACTION (2.294 Å) | | Cite: | Crystal structure of native Ara127N, a GH127 beta-L-arabinofuranosidase from Geobacillus Stearothermophilus T6
To be Published
|
|
4QK0
 
 | | Crystal structure of Ara127N-Se, a GH127 beta-L-arabinofuranosidase from Geobacillus Stearothermophilus T6 | | Descriptor: | GH127 beta-L-arabinofuranoside | | Authors: | Lansky, S, Salama, R, Dann, R, Shner, I, Manjasetty, B, Belrhali, H, Shoham, Y, Shoham, G. | | Deposit date: | 2014-06-05 | | Release date: | 2015-06-10 | | Method: | X-RAY DIFFRACTION (2.258 Å) | | Cite: | Crystal structure of Ara127N-Se, a GH127 beta-L-arabinofuranosidase from Geobacillus Stearothermophilus T6
To be Published
|
|
