6BOI
 
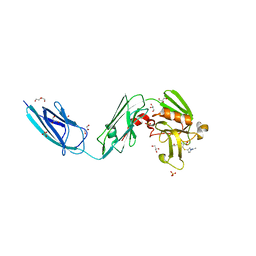 | | Crystal Structure of LdtMt2 (56-408) with a panipenem adduct at the active site cysteine-354 | | Descriptor: | (3S,5R)-5-[(2R,3R)-1,3-dihydroxybutan-2-yl]-3-({(3R)-1-[(1E)-ethanimidoyl]pyrrolidin-3-yl}sulfanyl)-L-proline, DI(HYDROXYETHYL)ETHER, GLYCEROL, ... | | Authors: | Saavedra, H, Bianchet, M.A. | | Deposit date: | 2017-11-20 | | Release date: | 2018-04-11 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (2.102 Å) | | Cite: | Structures and Mechanism of Inhibition of Mycobacterium tuberculosis L,D-transpeptidase 2 by Panipenem
To Be Published
|
|
2QHL
 
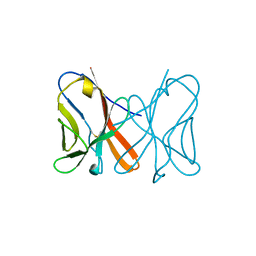 | | Crystal Structure of Novel Immune-Type Receptor 10 Extracellular Fragment from Ictalurus punctatus | | Descriptor: | Novel immune-type receptor 10 | | Authors: | Ostrov, D.A, Hernandez Prada, J.A, Haire, R.N, Cannon, J.P, Magis, A.T, Bailey, K.M, Litman, G.W. | | Deposit date: | 2007-07-02 | | Release date: | 2008-06-24 | | Last modified: | 2017-10-18 | | Method: | X-RAY DIFFRACTION (1.56 Å) | | Cite: | A bony fish immunological receptor of the NITR multigene family mediates allogeneic recognition.
Immunity, 29, 2008
|
|
2QJD
 
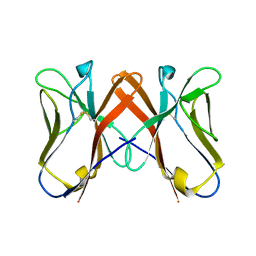 | | Crystal Structure of Novel Immune-Type Receptor 10 Extracellular Fragment Mutant N30D | | Descriptor: | Novel immune-type receptor 10 | | Authors: | Ostrov, D.A, Hernandez Prada, J.A, Haire, R.N, Cannon, J.P, Magis, A.T, Bailey, K.M, Litman, G.W. | | Deposit date: | 2007-07-06 | | Release date: | 2008-06-17 | | Last modified: | 2021-10-20 | | Method: | X-RAY DIFFRACTION (2.44 Å) | | Cite: | A bony fish immunological receptor of the NITR multigene family mediates allogeneic recognition.
Immunity, 29, 2008
|
|
2QTE
 
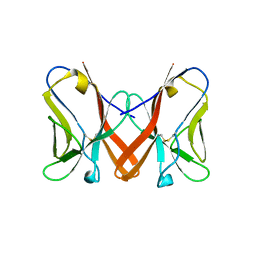 | | Crystal Structure of Novel Immune-Type Receptor 11 Extracellular Fragment Mutant N30D | | Descriptor: | Novel immune-type receptor 11 | | Authors: | Ostrov, D.A, Hernandez Prada, J.A, Haire, R.N, Cannon, J.P, Magis, A.T, Bailey, K.M, Litman, G.W. | | Deposit date: | 2007-08-01 | | Release date: | 2008-09-02 | | Last modified: | 2021-10-20 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | A bony fish immunological receptor of the NITR multigene family mediates allogeneic recognition.
Immunity, 29, 2008
|
|
2QQQ
 
 | | Crystal Structure of Novel Immune-Type Receptor 11 Extracellular Fragment from Ictalurus punctatus | | Descriptor: | Novel immune-type receptor 11 | | Authors: | Ostrov, D.A, Hernandez Prada, J.A, Haire, R.N, Cannon, J.P, Magis, A.T, Bailey, K.M, Litman, G.W. | | Deposit date: | 2007-07-26 | | Release date: | 2008-06-10 | | Last modified: | 2017-10-25 | | Method: | X-RAY DIFFRACTION (1.98 Å) | | Cite: | A bony fish immunological receptor of the NITR multigene family mediates allogeneic recognition.
Immunity, 29, 2008
|
|
