1BJB
 
 | | SOLUTION NMR STRUCTURE OF AMYLOID BETA[E16], RESIDUES 1-28, 14 STRUCTURES | | Descriptor: | AMYLOID BETA-PEPTIDE | | Authors: | Poulsen, S.-A, Watson, A.A, Craik, D.J. | | Deposit date: | 1998-06-23 | | Release date: | 1998-11-04 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | Solution structures in aqueous SDS micelles of two amyloid beta peptides of A beta(1-28) mutated at the alpha-secretase cleavage site (K16E, K16F)
J.Struct.Biol., 130, 2000
|
|
1CNN
 
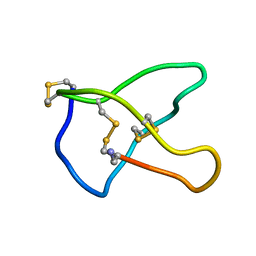 | | OMEGA-CONOTOXIN MVIIC FROM CONUS MAGUS | | Descriptor: | OMEGA-CONOTOXIN MVIIC | | Authors: | Nielsen, K.J, Adams, D, Thomas, L, Bond, T, Alewood, P.F, Craik, D.J, Lewis, R.J. | | Deposit date: | 1999-05-20 | | Release date: | 2000-05-31 | | Last modified: | 2023-12-27 | | Method: | SOLUTION NMR | | Cite: | Structure-activity relationships of omega-conotoxins MVIIA, MVIIC and 14 loop splice hybrids at N and P/Q-type calcium channels.
J.Mol.Biol., 289, 1999
|
|
2MSO
 
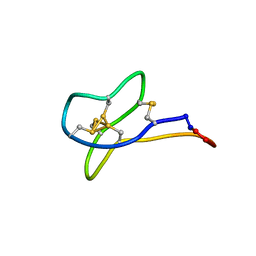 | | Solution study of cGm9a | | Descriptor: | Conotoxin Gm9.1 | | Authors: | Akcan, M, Clark, R.J, Daly, N.L, Conibear, A.C, de Faoite, A.C, Heghinian, M.C, Adams, D.J, Mari, F, Craik, D.J. | | Deposit date: | 2014-08-05 | | Release date: | 2015-07-22 | | Last modified: | 2023-12-27 | | Method: | SOLUTION NMR | | Cite: | Transforming conotoxins into cyclotides: Backbone cyclization of P-superfamily conotoxins.
Biopolymers, 104, 2015
|
|
1ONT
 
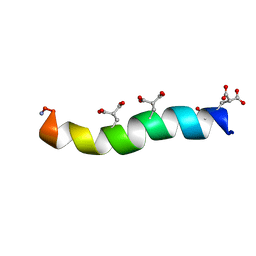 | | NMDA RECEPTOR ANTAGONIST, CONANTOKIN-T, NMR, 17 STRUCTURES | | Descriptor: | CONANTOKIN-T | | Authors: | Skjaerbaek, N, Nielsen, K.J, Lewis, R.J, Alewood, P.F, Craik, D.J. | | Deposit date: | 1996-08-27 | | Release date: | 1997-09-04 | | Last modified: | 2022-02-23 | | Method: | SOLUTION NMR | | Cite: | Determination of the solution structures of conantokin-G and conantokin-T by CD and NMR spectroscopy.
J.Biol.Chem., 272, 1997
|
|
1O8Z
 
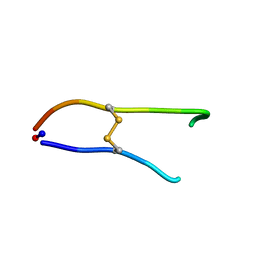 | | Solution structure of SFTI-1(6,5), an acyclic permutant of the proteinase inhibitor SFTI-1, cis-trans-trans conformer (ct-A) | | Descriptor: | CYCLIC TRYPSIN INHIBITOR | | Authors: | Marx, U.C, Craik, D.J. | | Deposit date: | 2002-12-09 | | Release date: | 2003-03-13 | | Last modified: | 2011-07-13 | | Method: | SOLUTION NMR | | Cite: | Enzymatic Cyclization of a Potent Bowman-Birk Protease Inhibitor, Sunflower Trypsin Inhibitor-1, and Solution Structure of an Acyclic Precursor Peptide
J.Biol.Chem., 278, 2003
|
|
1ORX
 
 | | Solution Structure of the acyclic permutant des-(24-28)-kalata B1. | | Descriptor: | kalata B1 | | Authors: | Barry, D.G, Daly, N.L, Clark, R.J, Sando, L, Craik, D.J. | | Deposit date: | 2003-03-17 | | Release date: | 2003-06-24 | | Last modified: | 2024-10-09 | | Method: | SOLUTION NMR | | Cite: | Linearization of a naturally occurring circular protein maintains structure but eliminates hemolytic activity
Biochemistry, 42, 2003
|
|
1NB1
 
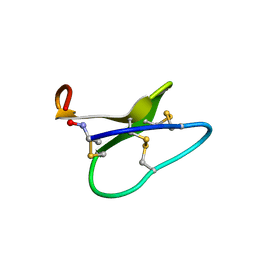 | | High resolution solution structure of kalata B1 | | Descriptor: | kalata B1 | | Authors: | Rosengren, K.J, Daly, N.L, Plan, M.R, Waine, C, Craik, D.J. | | Deposit date: | 2002-12-01 | | Release date: | 2003-03-18 | | Last modified: | 2024-10-23 | | Method: | SOLUTION NMR | | Cite: | Twists, Knots, and Rings in Proteins. STRUCTURAL DEFINITION OF THE CYCLOTIDE FRAMEWORK
J.Biol.Chem., 278, 2003
|
|
1O8Y
 
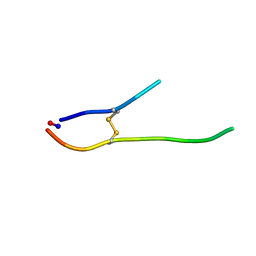 | | Solution structure of SFTI-1(6,5), an acyclic permutant of the proteinase inhibitor SFTI-1, trans-trans-trans conformer (tt-A) | | Descriptor: | CYCLIC TRYPSIN INHIBITOR | | Authors: | Marx, U.C, Craik, D.J. | | Deposit date: | 2002-12-09 | | Release date: | 2003-03-13 | | Last modified: | 2024-10-16 | | Method: | SOLUTION NMR | | Cite: | Enzymatic Cyclization of a Potent Bowman-Birk Protease Inhibitor, Sunflower Trypsin Inhibitor-1, and Solution Structure of an Acyclic Precursor Peptide
J.Biol.Chem., 278, 2003
|
|
1PEN
 
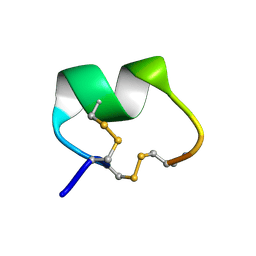 | | ALPHA-CONOTOXIN PNI1 | | Descriptor: | ALPHA-CONOTOXIN PNIA | | Authors: | Hu, S.-H, Gehrmann, J, Guddat, L.W, Alewood, P.F, Craik, D.J, Martin, J.L. | | Deposit date: | 1996-01-29 | | Release date: | 1997-04-21 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (1.1 Å) | | Cite: | The 1.1 A crystal structure of the neuronal acetylcholine receptor antagonist, alpha-conotoxin PnIA from Conus pennaceus.
Structure, 4, 1996
|
|
1ONU
 
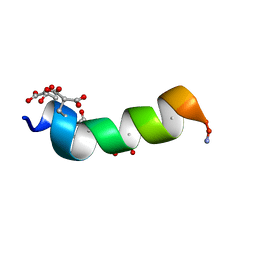 | | NMDA RECEPTOR ANTAGONIST, CONANTOKIN-G, NMR, 17 STRUCTURES | | Descriptor: | CONANTOKIN-G | | Authors: | Skjaerbaek, N, Nielsen, K.J, Lewis, R.J, Alewood, P.F, Craik, D.J. | | Deposit date: | 1996-08-27 | | Release date: | 1997-09-04 | | Last modified: | 2022-02-23 | | Method: | SOLUTION NMR | | Cite: | Determination of the solution structures of conantokin-G and conantokin-T by CD and NMR spectroscopy.
J.Biol.Chem., 272, 1997
|
|
1MII
 
 | | SOLUTION STRUCTURE OF ALPHA-CONOTOXIN MII | | Descriptor: | PROTEIN (ALPHA CONOTOXIN MII) | | Authors: | Hill, J.M, Oomen, C.J, Miranda, L.P, Bingham, J.P, Alewood, P.F, Craik, D.J. | | Deposit date: | 1998-10-05 | | Release date: | 1998-10-21 | | Last modified: | 2022-02-23 | | Method: | SOLUTION NMR | | Cite: | Three-dimensional solution structure of alpha-conotoxin MII by NMR spectroscopy: effects of solution environment on helicity.
Biochemistry, 37, 1998
|
|
1NBJ
 
 | | High-resolution solution structure of cycloviolacin O1 | | Descriptor: | cycloviolacin O1 | | Authors: | Rosengren, K.J, Daly, N.L, Plan, M.R, Waine, C, Craik, D.J. | | Deposit date: | 2002-12-02 | | Release date: | 2003-03-18 | | Last modified: | 2024-10-23 | | Method: | SOLUTION NMR | | Cite: | Twists, Knots, and Rings in Proteins. STRUCTURAL DEFINITION OF THE CYCLOTIDE FRAMEWORK.
J.Biol.Chem., 278, 2003
|
|
1MR4
 
 | | Solution Structure of NaD1 from Nicotiana alata | | Descriptor: | Nicotiana alata plant defensin 1 (NaD1) | | Authors: | Lay, F.T, Schirra, H.J, Scanlon, M.J, Anderson, M.A, Craik, D.J. | | Deposit date: | 2002-09-18 | | Release date: | 2003-09-18 | | Last modified: | 2024-10-23 | | Method: | SOLUTION NMR | | Cite: | The Three-dimensional Solution Structure of NaD1, a New Floral Defensin from Nicotiana alata and its Application to a Homology Model of the Crop Defense Protein alfAFP
J.MOL.BIOL., 325, 2003
|
|
1PT4
 
 | | Solution structure of the Moebius cyclotide kalata B2 | | Descriptor: | kalata B2 | | Authors: | Jennings, C.V, Anderson, M.A, Daly, N.L, Rosengren, K.J, Craik, D.J. | | Deposit date: | 2003-06-23 | | Release date: | 2004-08-10 | | Last modified: | 2024-10-30 | | Method: | SOLUTION NMR | | Cite: | Isolation, Solution Structure, and Insecticidal Activity of Kalata B2, a Circular Protein with a Twist: Do Mobius Strips Exist in Nature?(,)
Biochemistry, 44, 2005
|
|
1Q8L
 
 | | Second Metal Binding Domain of the Menkes ATPase | | Descriptor: | Copper-transporting ATPase 1 | | Authors: | Jones, C.E, Daly, N.L, Cobine, P.A, Craik, D.J, Dameron, C.T. | | Deposit date: | 2003-08-21 | | Release date: | 2004-01-20 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | Structure and metal binding studies of the second copper binding domain of the Menkes ATPase.
J.Struct.Biol., 143, 2003
|
|
1MVI
 
 | | N-TYPE CALCIUM CHANNEL BLOCKER, OMEGA-CONOTOXIN MVIIA, NMR, 15 STRUCTURES | | Descriptor: | MVIIA | | Authors: | Nielsen, K.J, Thomas, L, Lewis, R.J, Alewood, P.F, Craik, D.J. | | Deposit date: | 1996-08-02 | | Release date: | 1997-08-12 | | Last modified: | 2017-11-29 | | Method: | SOLUTION NMR | | Cite: | A consensus structure for omega-conotoxins with different selectivities for voltage-sensitive calcium channel subtypes: comparison of MVIIA, SVIB and SNX-202.
J.Mol.Biol., 263, 1996
|
|
1N4N
 
 | | Structure of the Plant Defensin PhD1 from Petunia Hybrida | | Descriptor: | floral defensin-like protein 1 | | Authors: | Janssen, B.J.C, Schirra, H.J, Lay, F.T, Anderson, M.A, Craik, D.J. | | Deposit date: | 2002-11-01 | | Release date: | 2003-11-01 | | Last modified: | 2024-10-16 | | Method: | SOLUTION NMR | | Cite: | Structure of Petunia hybrida defensin 1, a novel plant defensin with five disulfide bonds
Biochemistry, 42, 2003
|
|
1N1U
 
 | | NMR structure of [Ala1,15]kalata B1 | | Descriptor: | kalata B1 | | Authors: | Daly, N.L, Clark, R.J, Craik, D.J. | | Deposit date: | 2002-10-20 | | Release date: | 2003-02-25 | | Last modified: | 2024-10-16 | | Method: | SOLUTION NMR | | Cite: | Disulfide Folding Pathways of Cystine Knot Proteins. TYING THE KNOT WITHIN THE CIRCULAR BACKBONE OF THE CYCLOTIDES
J.Biol.Chem., 278, 2003
|
|
1MVJ
 
 | | N-TYPE CALCIUM CHANNEL BLOCKER, OMEGA-CONOTOXIN MVIIA NMR, 15 STRUCTURES | | Descriptor: | SVIB | | Authors: | Nielsen, K.J, Thomas, L, Lewis, R.J, Alewood, P.F, Craik, D.J. | | Deposit date: | 1996-08-02 | | Release date: | 1997-08-12 | | Last modified: | 2017-11-29 | | Method: | SOLUTION NMR | | Cite: | A consensus structure for omega-conotoxins with different selectivities for voltage-sensitive calcium channel subtypes: comparison of MVIIA, SVIB and SNX-202.
J.Mol.Biol., 263, 1996
|
|
1Q71
 
 | | The structure of microcin J25 is a threaded sidechain-to-backbone ring structure and not a head-to-tail cyclized backbone | | Descriptor: | microcin J25 | | Authors: | Rosengren, K.J, Clark, R, Daly, N.L, Goransson, U, Jones, A, Craik, D.J. | | Deposit date: | 2003-08-14 | | Release date: | 2003-12-16 | | Last modified: | 2012-12-12 | | Method: | SOLUTION NMR | | Cite: | Microcin J25 has a threaded sidechain-to-backbone ring structure and not a head-to-tail cyclized backbone.
J.Am.Chem.Soc., 125, 2003
|
|
1CNL
 
 | | ALPHA-CONOTOXIN IMI | | Descriptor: | PROTEIN (ALPHA-CONOTOXIN IMI) | | Authors: | Gehrmann, J, Daly, N.L, Craik, D.J. | | Deposit date: | 1999-05-20 | | Release date: | 1999-05-27 | | Last modified: | 2023-12-27 | | Method: | SOLUTION NMR | | Cite: | Solution structure of alpha-conotoxin ImI by 1H nuclear magnetic resonance.
J.Med.Chem., 42, 1999
|
|
1CE3
 
 | |
2NAW
 
 | |
2NAV
 
 | | NMR solution structure of Ex-4[1-16]/pl14a | | Descriptor: | Exendin-4, Alpha/kappa-conotoxin pl14a chimera | | Authors: | Schroeder, C.I, Swedberg, J.E, Craik, D.J. | | Deposit date: | 2016-01-11 | | Release date: | 2016-05-04 | | Last modified: | 2016-08-17 | | Method: | SOLUTION NMR | | Cite: | Truncated Glucagon-like Peptide-1 and Exendin-4 alpha-Conotoxin pl14a Peptide Chimeras Maintain Potency and alpha-Helicity and Reveal Interactions Vital for cAMP Signaling in Vitro.
J.Biol.Chem., 291, 2016
|
|
1GIB
 
 | | MU-CONOTOXIN GIIIB, NMR | | Descriptor: | MU-CONOTOXIN GIIIB | | Authors: | Hill, J.M, Alewood, P.F, Craik, D.J. | | Deposit date: | 1996-04-17 | | Release date: | 1996-11-08 | | Last modified: | 2022-02-23 | | Method: | SOLUTION NMR | | Cite: | Three-dimensional solution structure of mu-conotoxin GIIIB, a specific blocker of skeletal muscle sodium channels.
Biochemistry, 35, 1996
|
|
