5JOQ
 
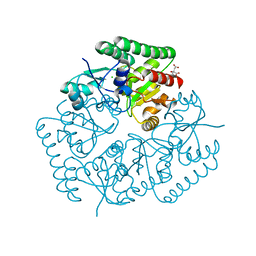 | | Crystal Structure of an ABC Transporter Substrate-Binding Protein from Listeria monocytogenes EGD-e | | Descriptor: | CHLORIDE ION, CITRIC ACID, Lmo2184 protein | | Authors: | Brunzelle, J.S, Wawrzak, Z, Kudritska, M, Savchenko, A, Anderson, W.F, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2016-05-02 | | Release date: | 2016-07-27 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (1.99 Å) | | Cite: | Crystal Structure of an ABC Transporter Substrate-Binding Protein from Listeria monocytogenes EGD-e
To Be Published
|
|
5JQC
 
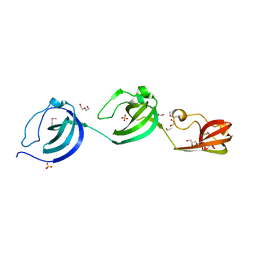 | | Crystal structure putative autolysin from Listeria monocytogenes | | Descriptor: | DI(HYDROXYETHYL)ETHER, GLYCEROL, Lmo1076 protein, ... | | Authors: | Chang, C, Zhou, M, Shatsman, S, Anderson, W.F, Joachimiak, A, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2016-05-04 | | Release date: | 2016-05-18 | | Last modified: | 2018-01-24 | | Method: | X-RAY DIFFRACTION (2.149 Å) | | Cite: | Crystal structure putative autolysin from Listeria monocytogenes
To Be Published
|
|
6X2D
 
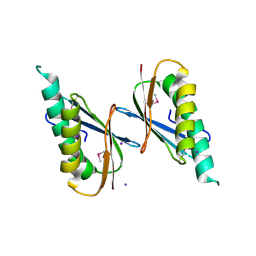 | | Crystal Structure of DNase I Domain of Ribonuclease E from Vibrio cholerae | | Descriptor: | IODIDE ION, Ribonuclease E | | Authors: | Minasov, G, Shuvalova, L, Kiryukhina, O, Dubrovska, I, Wiersum, G, Satchell, K.J.F, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2020-05-20 | | Release date: | 2020-06-10 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | Crystal Structure of DNase I Domain of Ribonuclease E from Vibrio cholerae.
To Be Published
|
|
5HW2
 
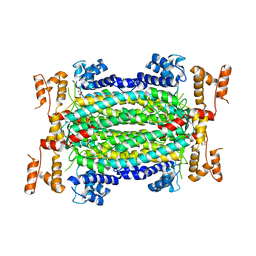 | | Crystal Structure of Adenylosuccinate Lyase from Francisella tularensis Complexed with fumaric acid | | Descriptor: | 1,2-ETHANEDIOL, Adenylosuccinate lyase, FUMARIC ACID, ... | | Authors: | Chang, C, Maltseva, N, Kim, Y, Shatsman, S, Anderson, W.F, Joachimiak, A, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2016-01-28 | | Release date: | 2016-02-10 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (2.054 Å) | | Cite: | Crystal Structure of Adenylosuccinate Lyase from Francisella tularensis Complexed with fumaric acid
To Be Published
|
|
5KKO
 
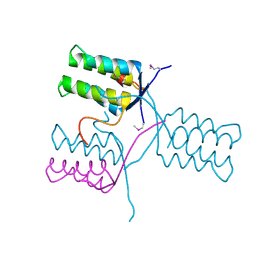 | | A 1.55A X-Ray Structure from Vibrio cholerae O1 biovar El Tor of a Hypothetical Protein | | Descriptor: | Uncharacterised protein | | Authors: | Brunzelle, J.S, Wawrzak, Z, Skarina, T, Savchenko, A, Anderson, W.F, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2016-06-22 | | Release date: | 2016-09-21 | | Method: | X-RAY DIFFRACTION (1.55 Å) | | Cite: | A 1.55A X-Ray Structure from Vibrio cholerae O1 biovar El Tor of a Hypothetical Protein
To Be Published
|
|
5HJ5
 
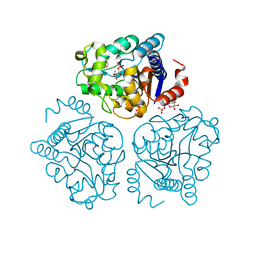 | | Crystal structure of tertiary complex of glucosamine-6-phosphate deaminase from Vibrio cholerae with BETA-D-GLUCOSE-6-PHOSPHATE and FRUCTOSE-6-PHOSPHATE | | Descriptor: | 6-O-phosphono-beta-D-glucopyranose, ACETIC ACID, FRUCTOSE -6-PHOSPHATE, ... | | Authors: | Chang, C, Maltseva, N, Kim, Y, Kwon, K, Anderson, W.F, Joachimiak, A, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2016-01-12 | | Release date: | 2016-02-03 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Crystal structure of tertiary complex of glucosamine-6-phosphate deaminase from Vibrio cholerae with BETA-D-GLUCOSE-6-PHOSPHATE and FRUCTOSE -6-PHOSPHATE
To Be Published
|
|
5KWS
 
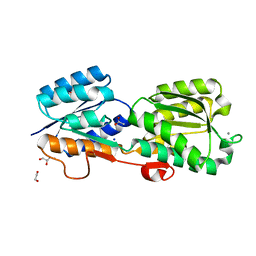 | | Crystal Structure of Galactose Binding Protein from Yersinia pestis in the Complex with beta D Glucose | | Descriptor: | 1,2-ETHANEDIOL, ACETIC ACID, CALCIUM ION, ... | | Authors: | Kim, Y, Maltseva, N, Mulligan, R, Grimshaw, S, Anderson, W.F, Joachimiak, A, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2016-07-19 | | Release date: | 2016-08-03 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (1.316 Å) | | Cite: | Crystal Structure of Galactose Binding Protein from Yersinia pestis in the Complex with beta D Glucose
To Be Published
|
|
6XJ3
 
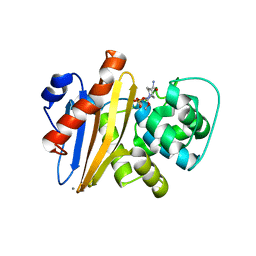 | | Crystal structure of Class D beta-lactamase from Klebsiella quasipneumoniae in complex with avibactam | | Descriptor: | (2S,5R)-1-formyl-5-[(sulfooxy)amino]piperidine-2-carboxamide, 1,2-ETHANEDIOL, 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, ... | | Authors: | Chang, C, Maltseva, N, Endres, M, Joachimiak, A, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2020-06-22 | | Release date: | 2020-07-01 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | Class D beta-lactamase from Klebsiella quasipneumoniae
To Be Published
|
|
8UFL
 
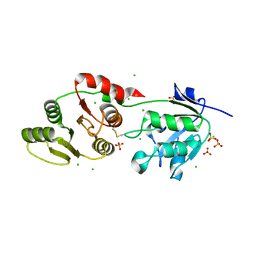 | | Crystal Structure of SARS-Unique Domain (SUD) of Nsp3 from SARS coronavirus | | Descriptor: | CHLORIDE ION, Papain-like protease nsp3, SULFATE ION | | Authors: | Minasov, G, Shuvalova, L, Rosas-Lemus, M, Kiryukhina, O, Brunzelle, J.S, Satchell, K.J.F, Center for Structural Genomics of Infectious Diseases (CSGID), Center for Structural Biology of Infectious Diseases (CSBID) | | Deposit date: | 2023-10-04 | | Release date: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (2.51 Å) | | Cite: | Crystal Structure of SARS-Unique Domain (SUD) of Nsp3 from SARS coronavirus
To Be Published
|
|
8UFM
 
 | | Crystal Structure of L516C/Y647C Mutant of SARS-Unique Domain (SUD) from SARS-CoV-2 | | Descriptor: | ACETATE ION, FORMIC ACID, Papain-like protease nsp3, ... | | Authors: | Minasov, G, Shuvalova, L, Brunzelle, J.S, Rosas-Lemus, M, Kiryukhina, O, Satchell, K.J.F, Center for Structural Genomics of Infectious Diseases (CSGID), Center for Structural Biology of Infectious Diseases (CSBID) | | Deposit date: | 2023-10-04 | | Release date: | 2023-10-18 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (1.65 Å) | | Cite: | Crystal Structure of L516C/Y647C Mutant of SARS-Unique Domain (SUD) from SARS-CoV-2
To Be Published
|
|
5JD8
 
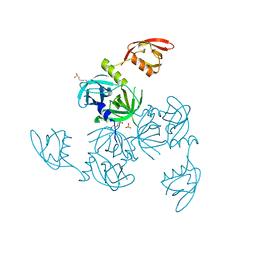 | | Crystal structure of the serine endoprotease from Yersinia pestis | | Descriptor: | 3-CYCLOHEXYL-1-PROPYLSULFONIC ACID, DI(HYDROXYETHYL)ETHER, Periplasmic serine peptidase DegS, ... | | Authors: | Filippova, E.V, Wawrzsak, Z, Sandoval, J, Skarina, T, Grimshaw, S, Savchenko, A, Anderson, W.F, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2016-04-15 | | Release date: | 2016-05-04 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | Crystal structure of the serine endoprotease from Yersinia pestis
To Be Published
|
|
8EY3
 
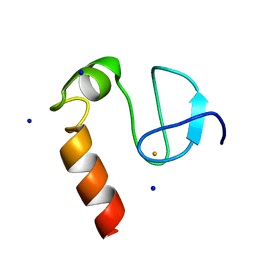 | | Contact-dependent growth inhibition (CDI) immunity protein from E. coli O32:H37 | | Descriptor: | Cys_rich_CPCC domain-containing protein, FE (III) ION, SODIUM ION | | Authors: | Michalska, K, Stols, L, Eschenfeldt, W, Goulding, C.W, Hayes, C.S, Joachimiak, A, Midwest Center for Structural Genomics (MCSG), Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2022-10-26 | | Release date: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (1 Å) | | Cite: | Contact-dependent growth inhibition (CDI) immunity protein from E. coli O32:H37
To Be Published
|
|
5HM8
 
 | | 2.85 Angstrom Crystal Structure of S-adenosylhomocysteinase from Cryptosporidium parvum in Complex with Adenosine and NAD. | | Descriptor: | ADENOSINE, Adenosylhomocysteinase, CHLORIDE ION, ... | | Authors: | Minasov, G, Shuvalova, L, Halavaty, A, Kiryukhina, O, Dubrovska, I, Bishop, B, Kwon, K, Anderson, W.F, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2016-01-15 | | Release date: | 2016-01-27 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (2.85 Å) | | Cite: | 2.85 Angstrom Crystal Structure of S-adenosylhomocysteinase from Cryptosporidium parvum in Complex with Adenosine and NAD.
To Be Published
|
|
5ICR
 
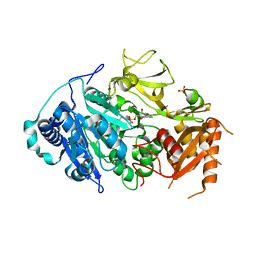 | | 2.25 Angstrom Resolution Crystal Structure of Fatty-Acid-CoA Ligase (FadD32) from Mycobacterium smegmatis in complex with Inhibitor 5'-O-[(11-phenoxyundecanoyl)sulfamoyl]adenosine. | | Descriptor: | 5'-O-[(11-phenoxyundecanoyl)sulfamoyl]adenosine, Acyl-CoA synthase, CHLORIDE ION, ... | | Authors: | Minasov, G, Shuvalova, L, Hung, D, Fisher, S.L, Edelstein, J, Kiryukhina, O, Dubrovska, I, Anderson, W.F, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2016-02-23 | | Release date: | 2016-04-06 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (2.25 Å) | | Cite: | 2.25 Angstrom Resolution Crystal Structure of Fatty-Acid-CoA Ligase (FadD32) from Mycobacterium smegmatis in complex with Inhibitor 5'-O-[(11-phenoxyundecanoyl)sulfamoyl]adenosine.
To Be Published
|
|
5KVR
 
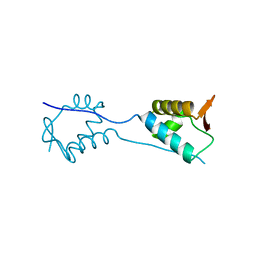 | | X-Ray Crystal Structure of a Fragment (1-75) of a Transcriptional Regulator PdhR from Escherichia coli CFT073 | | Descriptor: | Pyruvate dehydrogenase complex repressor | | Authors: | Brunzelle, J.S, Wawrzak, Z, Sandoval, J, Skarina, T, Savchenko, A, Anderson, W.F, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2016-07-15 | | Release date: | 2016-11-02 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (1.36 Å) | | Cite: | X-Ray Crystal Structure of a Fragment (1-75) of a Transcriptional Regulator PdhR from Escherichia coli CFT073
To Be Published
|
|
5J7R
 
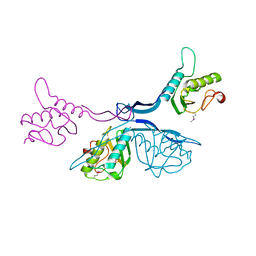 | | 2.5 Angstrom Crystal Structure of Putative Lipoprotein from Clostridium perfringens | | Descriptor: | CHLORIDE ION, DI(HYDROXYETHYL)ETHER, MAGNESIUM ION, ... | | Authors: | Minasov, G, Shuvalova, L, Kiryukhina, O, Flores, K, Shatsman, S, Kwon, K, Anderson, W.F, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2016-04-06 | | Release date: | 2016-04-20 | | Last modified: | 2016-05-04 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | 2.5 Angstrom Crystal Structure of Putative Lipoprotein from Clostridium perfringens
To Be Published
|
|
5JCV
 
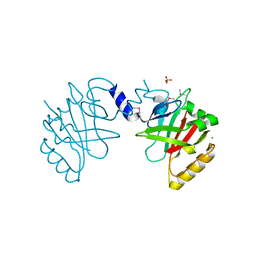 | | Sortase B from Listeria monocytogenes. | | Descriptor: | CHLORIDE ION, Lmo2181 protein, SULFATE ION | | Authors: | Osipiuk, J, Zhou, M, Grimshaw, S, Anderson, W.F, Joachimiak, A, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2016-04-15 | | Release date: | 2016-04-27 | | Last modified: | 2019-12-11 | | Method: | X-RAY DIFFRACTION (2.23 Å) | | Cite: | Sortase B from Listeria monocytogenes.
to be published
|
|
6MGR
 
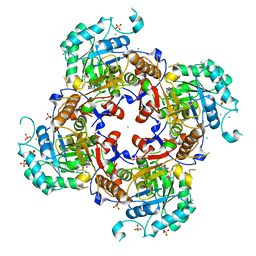 | | Crystal Structure of the Catalytic Domain of the Inosine Monophosphate Dehydrogenase from Campylobacter jejuni in the complex with inhibitor Oxanosine monophosphate | | Descriptor: | (4S)-2-METHYL-2,4-PENTANEDIOL, 5-[(Z)-(aminomethylidene)amino]-1-(5-O-phosphono-beta-D-ribofuranosyl)-1H-imidazole-4-carboxylic acid, CHLORIDE ION, ... | | Authors: | Kim, Y, Maltseva, N, Yu, R, Hedstrom, L, Joachimiak, A, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2018-09-14 | | Release date: | 2018-10-24 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (1.97 Å) | | Cite: | Crystal Structure of the Catalytic Domain of the Inosine Monophosphate Dehydrogenase from Campylobacter jejuni in the complex with inhibitor Oxanosine Monophosphate
To Be Published
|
|
6WY4
 
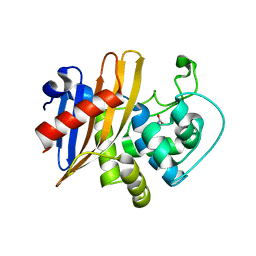 | | Crystal Structure of Wild Type Class D beta-lactamase from Clostridium difficile 630 | | Descriptor: | Beta-lactamase, DI(HYDROXYETHYL)ETHER, SODIUM ION | | Authors: | Minasov, G, Shuvalova, L, Dubrovska, I, Rosas-Lemus, M, Jedrzejczak, R, Satchell, K.J.F, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2020-05-12 | | Release date: | 2020-05-27 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Crystal Structure of Wild Type Class D beta-lactamase from Clostridium difficile 630
To Be Published
|
|
6WXC
 
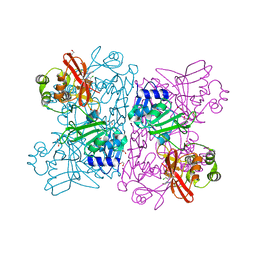 | | Crystal Structure of NSP15 Endoribonuclease from SARS CoV-2 in the Complex with potential repurposing drug Tipiracil | | Descriptor: | 1,2-ETHANEDIOL, 5-CHLORO-6-(1-(2-IMINOPYRROLIDINYL) METHYL) URACIL, FORMIC ACID, ... | | Authors: | Kim, Y, Maltseva, N, Jedrzejczak, R, Welk, L, Endres, M, Chang, C, Michalska, K, Joachimiak, A, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2020-05-10 | | Release date: | 2020-05-20 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | Tipiracil binds to uridine site and inhibits Nsp15 endoribonuclease NendoU from SARS-CoV-2.
Commun Biol, 4, 2021
|
|
6WLC
 
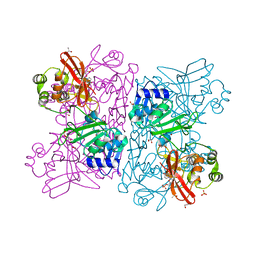 | | Crystal Structure of NSP15 Endoribonuclease from SARS CoV-2 in the Complex with Uridine-5'-Monophosphate | | Descriptor: | 1,2-ETHANEDIOL, 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, ACETATE ION, ... | | Authors: | Kim, Y, Maltseva, N, Jedrzejczak, R, Endres, M, Chang, C, Godzik, A, Michalska, K, Joachimiak, A, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2020-04-19 | | Release date: | 2020-04-29 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (1.82 Å) | | Cite: | Tipiracil binds to uridine site and inhibits Nsp15 endoribonuclease NendoU from SARS-CoV-2.
Commun Biol, 4, 2021
|
|
5K99
 
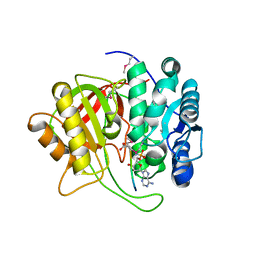 | | Crystal structure of microcin immunity protein MccF from Bacillus anthracis in complex with McC | | Descriptor: | ADENOSINE MONOPHOSPHATE, Microcin C, Microcin C7 self-immunity protein mccF | | Authors: | Nocek, B, Kulikovsky, A, Severinov, K, Dubiley, S, Joachimiak, A, Anderson, W.F, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2016-05-31 | | Release date: | 2017-03-22 | | Last modified: | 2024-05-15 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Crystal structure of microcin immunity protein MccF from Bacillus anthracis in complex with McC
To Be Published
|
|
5KJP
 
 | |
5HX7
 
 | | Metal ABC transporter from Listeria monocytogenes | | Descriptor: | Manganese-binding lipoprotein MntA | | Authors: | Osipiuk, J, Zhou, M, Grimshaw, S, Anderson, W.F, Joachimiak, A, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2016-01-29 | | Release date: | 2016-02-17 | | Last modified: | 2019-12-11 | | Method: | X-RAY DIFFRACTION (1.75 Å) | | Cite: | Metal ABC transporter from Listeria monocytogenes
to be published
|
|
5HSF
 
 | | 1.52 Angstrom Crystal Structure of Fc fragment of Human IgG1. | | Descriptor: | Ig gamma-1 chain C region, TRIETHYLENE GLYCOL | | Authors: | Minasov, G, Halavaty, A, Shuvalova, L, Dubrovska, I, Winsor, J, Flores, K, Bishop, B, Kwon, K, Anderson, W.F, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2016-01-25 | | Release date: | 2016-02-03 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (1.52 Å) | | Cite: | 1.52 Angstrom Crystal Structure of Fc fragment of Human IgG1.
To Be Published
|
|
