7FGU
 
 | |
3WFT
 
 | | Crystal structure of horse heart myoglobin reconstituted with cobalt(II) tetradehydrocorrin | | Descriptor: | (1R,19R) cobalt tetradehydrocorrin, (1S,19S) cobalt tetradehydrocorrin, GLYCEROL, ... | | Authors: | Mizohata, E, Morita, Y, Oohora, K, Hirata, K, Ohbayashi, J, Inoue, T, Hisaeda, Y, Hayashi, T. | | Deposit date: | 2013-07-23 | | Release date: | 2014-12-03 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (1.3 Å) | | Cite: | Co(II)/Co(I) reduction-induced axial histidine-flipping in myoglobin reconstituted with a cobalt tetradehydrocorrin as a methionine synthase model.
Chem.Commun.(Camb.), 50, 2014
|
|
3WJ4
 
 | | Crystal structure of PPARgamma ligand binding domain in complex with tributyltin | | Descriptor: | Peroxisome proliferator-activated receptor gamma, tributylstannanyl | | Authors: | Harada, S, Hiromori, Y, Fukakusa, S, Kawahara, K, Nakamura, S, Noda, M, Uchiyama, S, Fukui, K, Nishikawa, J, Nagase, H, Kobayashi, Y, Ohkubo, T, Yoshida, T, Nakanishi, T. | | Deposit date: | 2013-10-04 | | Release date: | 2014-10-15 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | Structural basis for PPARgamma transactivation by endocrine disrupting organotin compounds
To be Published
|
|
3WJ5
 
 | | Crystal structure of PPARgamma ligand binding domain in complex with triphenyltin | | Descriptor: | Peroxisome proliferator-activated receptor gamma, triphenylstannanyl | | Authors: | Harada, S, Hiromori, Y, Fukakusa, S, Kawahara, K, Nakamura, S, Noda, M, Uchiyama, S, Fukui, K, Nishikawa, J, Nagase, H, Kobayashi, Y, Ohkubo, T, Yoshida, T, Nakanishi, T. | | Deposit date: | 2013-10-04 | | Release date: | 2014-10-15 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (1.89 Å) | | Cite: | Structural basis for PPARgamma transactivation by endocrine disrupting organotin compounds
To be Published
|
|
3WFU
 
 | | Crystal structure of horse heart myoglobin reconstituted with cobalt(I) tetradehydrocorrin | | Descriptor: | (1R,19R) cobalt tetradehydrocorrin, (1S,19S) cobalt tetradehydrocorrin, Myoglobin, ... | | Authors: | Mizohata, E, Morita, Y, Oohora, K, Hirata, K, Ohbayashi, J, Inoue, T, Hisaeda, Y, Hayashi, T. | | Deposit date: | 2013-07-23 | | Release date: | 2014-12-03 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (1.35 Å) | | Cite: | Co(II)/Co(I) reduction-induced axial histidine-flipping in myoglobin reconstituted with a cobalt tetradehydrocorrin as a methionine synthase model.
Chem.Commun.(Camb.), 50, 2014
|
|
3WCQ
 
 | | Crystal structure analysis of Cyanidioschyzon melorae ferredoxin D58N mutant | | Descriptor: | FE2/S2 (INORGANIC) CLUSTER, Ferredoxin | | Authors: | Ueno, Y, Matsumoto, T, Yamano, A, Imai, T, Morimoto, Y. | | Deposit date: | 2013-05-31 | | Release date: | 2013-08-07 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (0.97 Å) | | Cite: | Increasing the electron-transfer ability of Cyanidioschyzon merolae ferredoxin by a one-point mutation - A high resolution and Fe-SAD phasing crystal structure analysis of the Asp58Asn mutant
Biochem.Biophys.Res.Commun., 436, 2013
|
|
5YXE
 
 | | Crystal Structure Analysis of feline serum albumin | | Descriptor: | Serum albumin | | Authors: | Kihira, K, Yokomaku, K, Akiyama, M, Morita, Y, Komatsu, T. | | Deposit date: | 2017-12-05 | | Release date: | 2018-04-04 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (3.40241528 Å) | | Cite: | Core-shell protein cluster comprising haemoglobin and recombinant feline serum albumin as an artificial O2 carrier for cats
J Mater Chem B., 2018
|
|
3WCU
 
 | | The structure of a deoxygenated 400 kda hemoglobin provides a more accurate description of the cooperative mechanism of giant hemoglobins: Deoxygenated form | | Descriptor: | A1 globin chain of giant V2 hemoglobin, A2 globin chain of giant V2 hemoglobin, B1 globin chain of giant V2 hemoglobin, ... | | Authors: | Numoto, N, Nakagawa, T, Ohara, R, Hasegawa, T, Kita, A, Yoshida, T, Maruyama, T, Imai, K, Fukumori, Y, Miki, K. | | Deposit date: | 2013-06-01 | | Release date: | 2014-06-04 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.9 Å) | | Cite: | The structure of a deoxygenated 400 kDa haemoglobin reveals ternary- and quaternary-structural changes of giant haemoglobins
Acta Crystallogr.,Sect.D, 70, 2014
|
|
3WCV
 
 | | The structure of a deoxygenated 400 kda hemoglobin provides a more accurate description of the cooperative mechanism of giant hemoglobins: CA bound form | | Descriptor: | A1 globin chain of giant V2 hemoglobin, A2 globin chain of giant V2 hemoglobin, B1 globin chain of giant V2 hemoglobin, ... | | Authors: | Numoto, N, Nakagawa, T, Ohara, R, Hasegawa, T, Kita, A, Yoshida, T, Maruyama, T, Imai, K, Fukumori, Y, Miki, K. | | Deposit date: | 2013-06-01 | | Release date: | 2014-06-04 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | The structure of a deoxygenated 400 kDa haemoglobin reveals ternary- and quaternary-structural changes of giant haemoglobins
Acta Crystallogr.,Sect.D, 70, 2014
|
|
3WMQ
 
 | | Crystal structure of the complex between SLL-2 and GalNAc. | | Descriptor: | (4S)-2-METHYL-2,4-PENTANEDIOL, 2-acetamido-2-deoxy-alpha-D-galactopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Kita, A, Jimbo, M, Sakai, R, Morimoto, Y, Miki, K. | | Deposit date: | 2013-11-22 | | Release date: | 2015-01-14 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Crystal structure of a symbiosis-related lectin from octocoral.
Glycobiology, 25, 2015
|
|
3WCW
 
 | | The structure of a deoxygenated 400 kda hemoglobin provides a more accurate description of the cooperative mechanism of giant hemoglobins: MG bound form | | Descriptor: | A1 globin chain of giant V2 hemoglobin, A2 globin chain of giant V2 hemoglobin, B1 globin chain of giant V2 hemoglobin, ... | | Authors: | Numoto, N, Nakagawa, T, Ohara, R, Hasegawa, T, Kita, A, Yoshida, T, Maruyama, T, Imai, K, Fukumori, Y, Miki, K. | | Deposit date: | 2013-06-01 | | Release date: | 2014-06-04 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | The structure of a deoxygenated 400 kDa haemoglobin reveals ternary- and quaternary-structural changes of giant haemoglobins
Acta Crystallogr.,Sect.D, 70, 2014
|
|
3WMP
 
 | | Crystal structure of SLL-2 | | Descriptor: | (4S)-2-METHYL-2,4-PENTANEDIOL, 2-acetamido-2-deoxy-alpha-D-galactopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Kita, A, Jimbo, M, Sakai, R, Morimoto, Y, Miki, K. | | Deposit date: | 2013-11-22 | | Release date: | 2015-01-14 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Crystal structure of a symbiosis-related lectin from octocoral.
Glycobiology, 25, 2015
|
|
3WCT
 
 | | The structure of a deoxygenated 400 kda hemoglobin provides a more accurate description of the cooperative mechanism of giant hemoglobins: Oxygenated form | | Descriptor: | A1 globin chain of giant V2 hemoglobin, A2 globin chain of giant V2 hemoglobin, B1 globin chain of giant V2 hemoglobin, ... | | Authors: | Numoto, N, Nakagawa, T, Ohara, R, Hasegawa, T, Kita, A, Yoshida, T, Maruyama, T, Imai, K, Fukumori, Y, Miki, K. | | Deposit date: | 2013-06-01 | | Release date: | 2014-06-04 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | The structure of a deoxygenated 400 kDa haemoglobin reveals ternary- and quaternary-structural changes of giant haemoglobins
Acta Crystallogr.,Sect.D, 70, 2014
|
|
8JU3
 
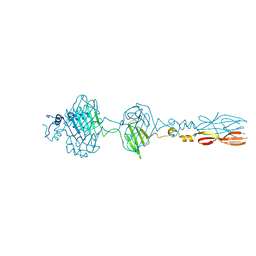 | | Mu phage tail fiber | | Descriptor: | Tail fiber protein S,Tail fiber protein S' | | Authors: | Yamashita, E, Takeda, S. | | Deposit date: | 2023-06-24 | | Release date: | 2024-02-28 | | Last modified: | 2024-09-11 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Determination of the three-dimensional structure of bacteriophage Mu(-) tail fiber and its characterization.
Virology, 593, 2024
|
|
3VYE
 
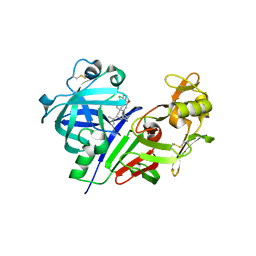 | | Human renin in complex with inhibitor 7 | | Descriptor: | (3S,5R)-5-[4-(2-chlorophenyl)-2,2-dimethyl-5-oxopiperazin-1-yl]-N-(3-methylbutyl)piperidine-3-carboxamide, 2-acetamido-2-deoxy-beta-D-glucopyranose, Renin | | Authors: | Takahashi, M, Matsui, Y, Hanzawa, H. | | Deposit date: | 2012-09-24 | | Release date: | 2012-12-19 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Design and discovery of new (3S,5R)-5-[4-(2-chlorophenyl)-2,2-dimethyl-5-oxopiperazin-1-yl]piperidine-3-carboxamides as potent renin inhibitors
Bioorg.Med.Chem.Lett., 22, 2012
|
|
3VYD
 
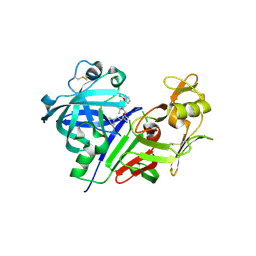 | | Human renin in complex with inhibitor 6 | | Descriptor: | (3S,5R)-5-{[4-(2-chlorophenyl)-2,2-dimethyl-5-oxopiperazin-1-yl]methyl}-N-(3-methylbutyl)piperidine-3-carboxamide, 2-acetamido-2-deoxy-beta-D-glucopyranose, Renin | | Authors: | Takahashi, M, Hanzawa, H. | | Deposit date: | 2012-09-24 | | Release date: | 2012-12-19 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.81 Å) | | Cite: | Design and discovery of new (3S,5R)-5-[4-(2-chlorophenyl)-2,2-dimethyl-5-oxopiperazin-1-yl]piperidine-3-carboxamides as potent renin inhibitors
Bioorg.Med.Chem.Lett., 22, 2012
|
|
3VYF
 
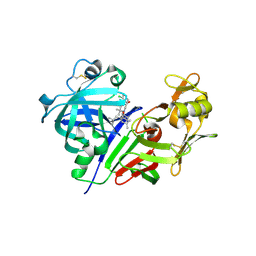 | | Human renin in complex with inhibitor 9 | | Descriptor: | (3S,5R)-5-[4-(2-chlorophenyl)-2,2-dimethyl-5-oxopiperazin-1-yl]-N-(2,6-dimethylheptan-4-yl)piperidine-3-carboxamide, 2-acetamido-2-deoxy-beta-D-glucopyranose, Renin | | Authors: | Takahashi, M, Hanzawa, H. | | Deposit date: | 2012-09-24 | | Release date: | 2012-12-19 | | Last modified: | 2024-10-09 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Design and discovery of new (3S,5R)-5-[4-(2-chlorophenyl)-2,2-dimethyl-5-oxopiperazin-1-yl]piperidine-3-carboxamides as potent renin inhibitors
Bioorg.Med.Chem.Lett., 22, 2012
|
|
6JU1
 
 | | p-Hydroxybenzoate hydroxylase Y385F mutant complexed with 3,4-dihydroxybenzoate | | Descriptor: | 3,4-DIHYDROXYBENZOIC ACID, 3,6,9,12,15,18-HEXAOXAICOSANE-1,20-DIOL, 4-hydroxybenzoate 3-monooxygenase, ... | | Authors: | Yato, M, Arakawa, T, Yamada, C, Fushinobu, S. | | Deposit date: | 2019-04-12 | | Release date: | 2019-11-06 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Understanding the Molecular Mechanism Underlying the High Catalytic Activity ofp-Hydroxybenzoate Hydroxylase Mutants for Producing Gallic Acid.
Biochemistry, 58, 2019
|
|
6KYW
 
 | | S8-mSRK-S8-SP11 complex | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, Receptor protein kinase SRK8, S locus protein 11 | | Authors: | Murase, K, Hakoshima, T, Mori, T. | | Deposit date: | 2019-09-20 | | Release date: | 2020-09-16 | | Last modified: | 2020-10-21 | | Method: | X-RAY DIFFRACTION (2.60112739 Å) | | Cite: | Mechanism of self/nonself-discrimination in Brassica self-incompatibility.
Nat Commun, 11, 2020
|
|
3FX5
 
 | | Structure of HIV-1 Protease in Complex with Potent Inhibitor KNI-272 Determined by High Resolution X-ray Crystallography | | Descriptor: | (4R)-N-tert-butyl-3-[(2S,3S)-2-hydroxy-3-({N-[(isoquinolin-5-yloxy)acetyl]-S-methyl-L-cysteinyl}amino)-4-phenylbutanoyl]-1,3-thiazolidine-4-carboxamide, GLYCEROL, protease | | Authors: | Adachi, M, Ohhara, T, Tamada, T, Okazaki, N, Kuroki, R. | | Deposit date: | 2009-01-20 | | Release date: | 2009-03-24 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (0.93 Å) | | Cite: | Structure of HIV-1 protease in complex with potent inhibitor KNI-272 determined by high-resolution X-ray and neutron crystallography.
Proc.Natl.Acad.Sci.USA, 2009
|
|
5GUX
 
 | | Cytochrome c-dependent nitric oxide reductase (cNOR) from Pseudomonas aeruginosa in complex with xenon | | Descriptor: | Antibody fab fragment heavy chain, Antibody fab fragment light chain, CALCIUM ION, ... | | Authors: | Ishii, S, Terasaka, E, Sugimoto, H, Shiro, Y, Tosha, T. | | Deposit date: | 2016-08-31 | | Release date: | 2017-08-16 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (3.3 Å) | | Cite: | Dynamics of nitric oxide controlled by protein complex in bacterial system.
Proc. Natl. Acad. Sci. U.S.A., 114, 2017
|
|
5GUW
 
 | | Complex of Cytochrome cd1 Nitrite Reductase and Nitric Oxide Reductase in Denitrification of Pseudomonas aeruginosa | | Descriptor: | CALCIUM ION, CHLORIDE ION, FE (III) ION, ... | | Authors: | Terasaka, E, Sugimoto, H, Shiro, Y, Tosha, T. | | Deposit date: | 2016-08-31 | | Release date: | 2017-08-16 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (3.2 Å) | | Cite: | Dynamics of nitric oxide controlled by protein complex in bacterial system
Proc. Natl. Acad. Sci. U.S.A., 114, 2017
|
|
3AQO
 
 | |
6L93
 
 | |
1V49
 
 | | Solution structure of microtubule-associated protein light chain-3 | | Descriptor: | Microtubule-associated proteins 1A/1B light chain 3B | | Authors: | Kouno, T, Mizuguchi, M, Tanida, I, Ueno, T, Kominami, E, Kawano, K. | | Deposit date: | 2003-11-11 | | Release date: | 2004-12-28 | | Last modified: | 2023-12-27 | | Method: | SOLUTION NMR | | Cite: | Solution structure of microtubule-associated protein light chain 3 and identification of its functional subdomains.
J.Biol.Chem., 280, 2005
|
|
