4E51
 
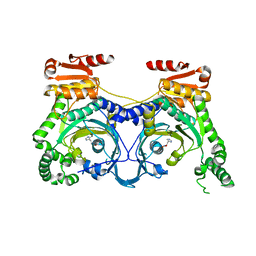 | |
4E17
 
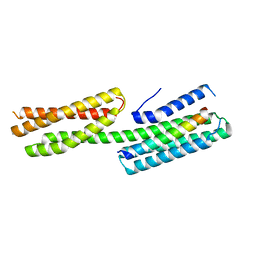 | | Alpha-E-catenin is an autoinhibited molecule that co-activates vinculin | | Descriptor: | Catenin alpha-1, Vinculin | | Authors: | Choi, H.-J, Pokutta, S, Cadwell, G.W, Bankston, L.A, Liddington, R.C, Weis, W.I. | | Deposit date: | 2012-03-05 | | Release date: | 2012-05-16 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (2.304 Å) | | Cite: | Conformational plasticity of alpha-catenin revealed by binding interactions with vinculin
To be Published
|
|
3EIY
 
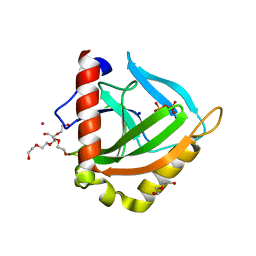 | |
3EQB
 
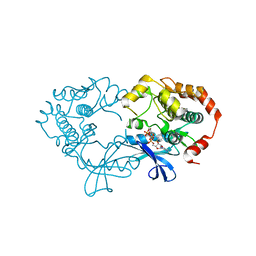 | | X-ray structure of the human mitogen-activated protein kinase kinase 1 (MEK1) in a complex with ligand and MgATP | | Descriptor: | ADENOSINE-5'-TRIPHOSPHATE, Dual specificity mitogen-activated protein kinase kinase 1, MAGNESIUM ION, ... | | Authors: | Ohren, J.F, Pavlovsky, A, Zhang, E. | | Deposit date: | 2008-09-30 | | Release date: | 2008-11-11 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (2.62 Å) | | Cite: | 2-Alkylamino- and alkoxy-substituted 2-amino-1,3,4-oxadiazoles-O-Alkyl benzohydroxamate esters replacements retain the desired inhibition and selectivity against MEK (MAP ERK kinase).
Bioorg.Med.Chem.Lett., 18, 2008
|
|
3F0D
 
 | |
3F0G
 
 | |
3F0E
 
 | |
3G2N
 
 | | Crystal structure of N-acylglucosylamine with glycogen phosphorylase | | Descriptor: | Glycogen phosphorylase, muscle form, N-(phenylcarbonyl)-beta-D-glucopyranosylamine | | Authors: | Chrysina, E.D, Bokor, E, Alexacou, K.-M, Charavgi, M.-D, Oikonomakos, G.N, Zographos, S.E, Leonidas, D.D, Oikonomakos, N.G, Somsak, L. | | Deposit date: | 2009-01-31 | | Release date: | 2010-02-16 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Amide-1,2,3-triazole bioisosterism: the glycogen phosphorylase case
Tetrahedron: Asymmetry, 20, 2009
|
|
3G2H
 
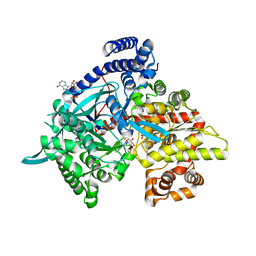 | | Crystal structure of 1-(beta-D-glucopyranosyl)-4-substituted-1,2,3-triazoles in complex with glycogen phosphorylase | | Descriptor: | 1-beta-D-glucopyranosyl-4-phenyl-1H-1,2,3-triazole, DIMETHYL SULFOXIDE, Glycogen phosphorylase, ... | | Authors: | Chrysina, E.D, Bokor, E, Alexacou, K.-M, Charavgi, M.-D, Oikonomakos, G.N, Zographos, S.E, Leonidas, D.D, Oikonomakos, N.G, Somsak, L. | | Deposit date: | 2009-01-31 | | Release date: | 2010-02-16 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.03 Å) | | Cite: | Amide-1,2,3-triazole bioisosterism: the glycogen phosphorylase case
Tetrahedron: Asymmetry, 20, 2009
|
|
3G2K
 
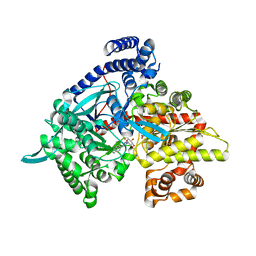 | | Crystal structure of 1-(beta-D-glucopyranosyl)-4-substituted-1,2,3-triazole | | Descriptor: | 1-beta-D-glucopyranosyl-4-naphthalen-2-yl-1H-1,2,3-triazole, Glycogen phosphorylase, muscle form | | Authors: | Chrysina, E.D, Bokor, E, Alexacou, K.-M, Charavgi, M.-D, Oikonomakos, G.N, Zographos, S.E, Leonidas, D.D, Oikonomakos, N.G, Somsak, L. | | Deposit date: | 2009-01-31 | | Release date: | 2010-02-16 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Amide-1,2,3-triazole bioisosterism: the glycogen phosphorylase case
Tetrahedron: Asymmetry, 20, 2009
|
|
3G2I
 
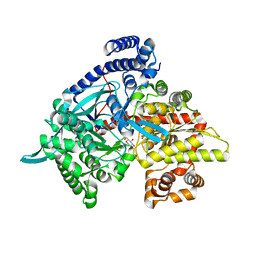 | | Crystal structure of 1-(beta-D-glucopyranosyl)-4-substituted-1,2,3-triazole | | Descriptor: | 1-beta-D-glucopyranosyl-4-(hydroxymethyl)-1H-1,2,3-triazole, Glycogen phosphorylase, muscle form | | Authors: | Chrysina, E.D, Bokor, E, Alexacou, K.-M, Charavgi, M.-D, Oikonomakos, G.N, Zographos, S.E, Leonidas, D.D, Oikonomakos, N.G, Somsak, L. | | Deposit date: | 2009-01-31 | | Release date: | 2010-02-16 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Amide-1,2,3-triazole bioisosterism: the glycogen phosphorylase case
Tetrahedron: Asymmetry, 20, 2009
|
|
3G2L
 
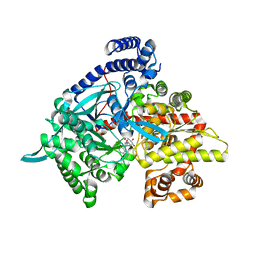 | | Crystal structure of 1-(beta-D-glucopyranosyl)-4-substituted-1,2,3-triazoles in complex with glycogen phosphorylase | | Descriptor: | 1-beta-D-glucopyranosyl-4-naphthalen-1-yl-1H-1,2,3-triazole, Glycogen phosphorylase, muscle form | | Authors: | Chrysina, E.D, Bokor, E, Alexacou, K.-M, Charavgi, M.-D, Oikonomakos, G.N, Zographos, S.E, Leonidas, D.D, Oikonomakos, N.G, Somsak, L. | | Deposit date: | 2009-01-31 | | Release date: | 2010-02-16 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Amide-1,2,3-triazole bioisosterism: the glycogen phosphorylase case
Tetrahedron: Asymmetry, 20, 2009
|
|
3G2J
 
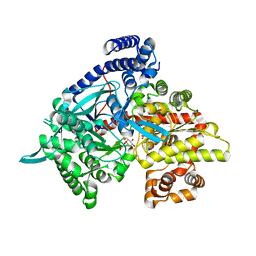 | | Crystal structure of 1-(beta-D-glucopyranosyl)-4-substituted-1,2,3-triazoles in complex with glycogen phosphorylase | | Descriptor: | Glycogen phosphorylase, muscle form, N-(hydroxyacetyl)-beta-D-glucopyranosylamine | | Authors: | Chrysina, E.D, Bokor, E, Alexacou, K.-M, Charavgi, M.-D, Oikonomakos, G.N, Zographos, S.E, Leonidas, D.D, Oikonomakos, N.G, Somsak, L. | | Deposit date: | 2009-01-31 | | Release date: | 2010-02-16 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.14 Å) | | Cite: | Amide-1,2,3-triazole bioisosterism: the glycogen phosphorylase case
Tetrahedron: Asymmetry, 20, 2009
|
|
3DMS
 
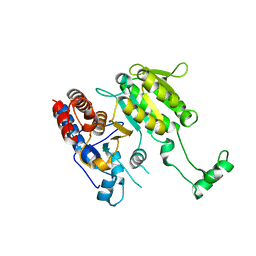 | |
3EJ0
 
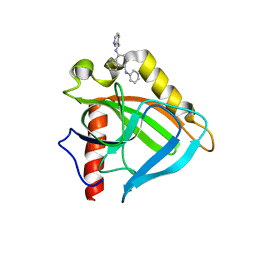 | |
3ERX
 
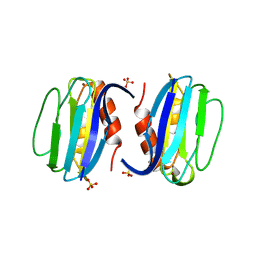 | | High-resolution structure of Paracoccus pantotrophus pseudoazurin | | Descriptor: | COPPER (II) ION, Pseudoazurin, SULFATE ION | | Authors: | Najmudin, S, Pauleta, S.R, Moura, I, Romao, M.J. | | Deposit date: | 2008-10-03 | | Release date: | 2009-10-13 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (1.25 Å) | | Cite: | The 1.4 A resolution structure of Paracoccus pantotrophus pseudoazurin.
Acta Crystallogr.,Sect.F, 66, 2010
|
|
3HGB
 
 | |
3NXS
 
 | |
3OE0
 
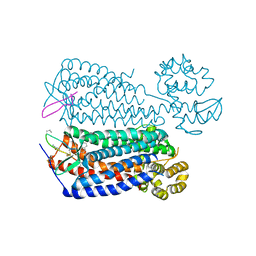 | | Crystal structure of the CXCR4 chemokine receptor in complex with a cyclic peptide antagonist CVX15 | | Descriptor: | C-X-C chemokine receptor type 4, Lysozyme Chimera, Polyphemusin analog, ... | | Authors: | Wu, B, Mol, C.D, Han, G.W, Katritch, V, Chien, E.Y.T, Liu, W, Cherezov, V, Stevens, R.C, Accelerated Technologies Center for Gene to 3D Structure (ATCG3D), GPCR Network (GPCR) | | Deposit date: | 2010-08-12 | | Release date: | 2010-10-27 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (2.9 Å) | | Cite: | Structures of the CXCR4 chemokine GPCR with small-molecule and cyclic peptide antagonists.
Science, 330, 2010
|
|
3OEC
 
 | |
3OE9
 
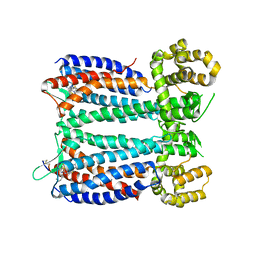 | | Crystal structure of the chemokine CXCR4 receptor in complex with a small molecule antagonist IT1t in P1 spacegroup | | Descriptor: | (6,6-dimethyl-5,6-dihydroimidazo[2,1-b][1,3]thiazol-3-yl)methyl N,N'-dicyclohexylimidothiocarbamate, C-X-C chemokine receptor type 4, Lysozyme Chimera | | Authors: | Wu, B, Mol, C.D, Han, G.W, Katritch, V, Chien, E.Y.T, Liu, W, Cherezov, V, Stevens, R.C, Accelerated Technologies Center for Gene to 3D Structure (ATCG3D), GPCR Network (GPCR) | | Deposit date: | 2010-08-12 | | Release date: | 2010-10-27 | | Last modified: | 2024-10-09 | | Method: | X-RAY DIFFRACTION (3.1 Å) | | Cite: | Structures of the CXCR4 chemokine GPCR with small-molecule and cyclic peptide antagonists.
Science, 330, 2010
|
|
3ODU
 
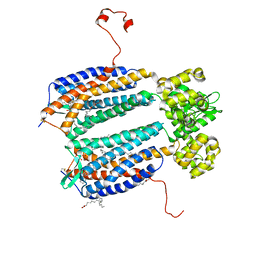 | | The 2.5 A structure of the CXCR4 chemokine receptor in complex with small molecule antagonist IT1t | | Descriptor: | (2R)-2,3-dihydroxypropyl (9Z)-octadec-9-enoate, (6,6-dimethyl-5,6-dihydroimidazo[2,1-b][1,3]thiazol-3-yl)methyl N,N'-dicyclohexylimidothiocarbamate, C-X-C chemokine receptor type 4, ... | | Authors: | Wu, B, Mol, C.D, Han, G.W, Katritch, V, Chien, E.Y.T, Liu, W, Cherezov, V, Stevens, R.C, Accelerated Technologies Center for Gene to 3D Structure (ATCG3D), GPCR Network (GPCR) | | Deposit date: | 2010-08-11 | | Release date: | 2010-10-27 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Structures of the CXCR4 chemokine GPCR with small-molecule and cyclic peptide antagonists.
Science, 330, 2010
|
|
3OE6
 
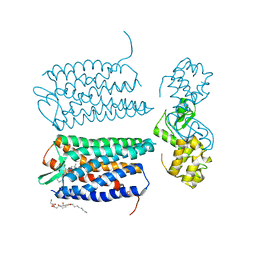 | | Crystal structure of the CXCR4 chemokine receptor in complex with a small molecule antagonist IT1t in I222 spacegroup | | Descriptor: | (2R)-2,3-dihydroxypropyl (9Z)-octadec-9-enoate, (6,6-dimethyl-5,6-dihydroimidazo[2,1-b][1,3]thiazol-3-yl)methyl N,N'-dicyclohexylimidothiocarbamate, C-X-C chemokine receptor type 4, ... | | Authors: | Wu, B, Mol, C.D, Han, G.W, Katritch, V, Chien, E.Y.T, Liu, W, Cherezov, V, Stevens, R.C, Accelerated Technologies Center for Gene to 3D Structure (ATCG3D), GPCR Network (GPCR) | | Deposit date: | 2010-08-12 | | Release date: | 2010-10-27 | | Last modified: | 2024-10-09 | | Method: | X-RAY DIFFRACTION (3.2 Å) | | Cite: | Structures of the CXCR4 chemokine GPCR with small-molecule and cyclic peptide antagonists.
Science, 330, 2010
|
|
3OE8
 
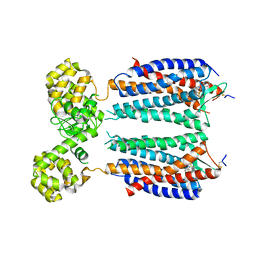 | | Crystal structure of the CXCR4 chemokine receptor in complex with a small molecule antagonist IT1t in P1 spacegroup | | Descriptor: | (6,6-dimethyl-5,6-dihydroimidazo[2,1-b][1,3]thiazol-3-yl)methyl N,N'-dicyclohexylimidothiocarbamate, C-X-C chemokine receptor type 4, Lysozyme Chimera | | Authors: | Wu, B, Mol, C.D, Han, G.W, Katritch, V, Chien, E.Y.T, Liu, W, Cherezov, V, Stevens, R.C, Accelerated Technologies Center for Gene to 3D Structure (ATCG3D), GPCR Network (GPCR) | | Deposit date: | 2010-08-12 | | Release date: | 2010-10-27 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (3.1 Å) | | Cite: | Structures of the CXCR4 chemokine GPCR with small-molecule and cyclic peptide antagonists.
Science, 330, 2010
|
|
3P10
 
 | |
