8GZV
 
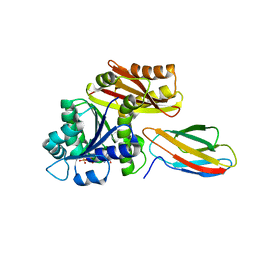 | | Klebsiella pneumoniae FtsZ complexed with monobody (P212121) | | Descriptor: | Cell division protein FtsZ, GUANOSINE-5'-DIPHOSPHATE, Monobody | | Authors: | Matsumura, H, Yoshizawa, T, Fujita, J, Tanaka, S, Amesaka, H. | | Deposit date: | 2022-09-27 | | Release date: | 2023-07-19 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Structures of a FtsZ single protofilament and a double-helical tube in complex with a monobody.
Nat Commun, 14, 2023
|
|
8GZX
 
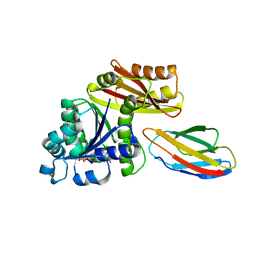 | | Escherichia coli FtsZ complexed with monobody (P212121) | | Descriptor: | Cell division protein FtsZ, GUANOSINE-5'-DIPHOSPHATE, Monobody | | Authors: | Matsumura, H, Yoshizawa, T, Fujita, J, Tanaka, S, Amesaka, H. | | Deposit date: | 2022-09-27 | | Release date: | 2023-07-19 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (1.84 Å) | | Cite: | Structures of a FtsZ single protofilament and a double-helical tube in complex with a monobody.
Nat Commun, 14, 2023
|
|
8GZW
 
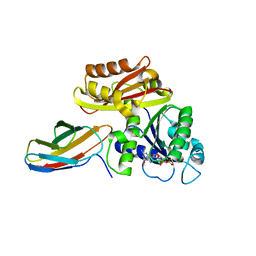 | | Klebsiella pneumoniae FtsZ complexed with monobody (P21) | | Descriptor: | Cell division protein FtsZ, GUANOSINE-5'-DIPHOSPHATE, Monobody | | Authors: | Matsumura, H, Yoshizawa, T, Fujita, J, Tanaka, S, Amesaka, H. | | Deposit date: | 2022-09-27 | | Release date: | 2023-07-19 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Structures of a FtsZ single protofilament and a double-helical tube in complex with a monobody.
Nat Commun, 14, 2023
|
|
8GZY
 
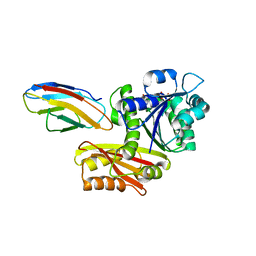 | | Escherichia coli FtsZ complexed with monobody (P21) | | Descriptor: | Cell division protein FtsZ, GUANOSINE-5'-DIPHOSPHATE, Monobody | | Authors: | Matsumura, H, Yoshizawa, T, Fujita, J, Tanaka, S, Amesaka, H. | | Deposit date: | 2022-09-27 | | Release date: | 2023-07-19 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Structures of a FtsZ single protofilament and a double-helical tube in complex with a monobody.
Nat Commun, 14, 2023
|
|
2E5V
 
 | | Crystal structure of L-Aspartate Oxidase from hyperthermophilic archaeon Sulfolobus tokodaii | | Descriptor: | CHLORIDE ION, FLAVIN-ADENINE DINUCLEOTIDE, L-aspartate oxidase | | Authors: | Yoneda, K, Sakuraba, H, Asai, I, Tsuge, H, Katunuma, N, Ohshima, T. | | Deposit date: | 2006-12-25 | | Release date: | 2008-01-01 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (2.09 Å) | | Cite: | Structure of l-aspartate oxidase from the hyperthermophilic archaeon Sulfolobus tokodaii
Biochim.Biophys.Acta, 1784, 2008
|
|
8HYE
 
 | | Structure of amino acid dehydrogenase-2752 with ligand | | Descriptor: | 1,2-ETHANEDIOL, 4-(2-HYDROXYETHYL)-1-PIPERAZINE ETHANESULFONIC ACID, Alanine dehydrogenase, ... | | Authors: | Sakuraba, H, Ohshima, T. | | Deposit date: | 2023-01-06 | | Release date: | 2023-04-05 | | Last modified: | 2024-05-29 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Two different alanine dehydrogenases from Geobacillus kaustophilus: Their biochemical characteristics and differential expression in vegetative cells and spores.
Biochim Biophys Acta Proteins Proteom, 1871, 2023
|
|
8HYH
 
 | | Structure of amino acid dehydrogenase3448 | | Descriptor: | 1,2-ETHANEDIOL, Alanine dehydrogenase, NICOTINAMIDE-ADENINE-DINUCLEOTIDE, ... | | Authors: | Sakuraba, H, Ohshima, T. | | Deposit date: | 2023-01-06 | | Release date: | 2023-04-05 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (2.39 Å) | | Cite: | Two different alanine dehydrogenases from Geobacillus kaustophilus: Their biochemical characteristics and differential expression in vegetative cells and spores.
Biochim Biophys Acta Proteins Proteom, 1871, 2023
|
|
3VNR
 
 | |
3WYB
 
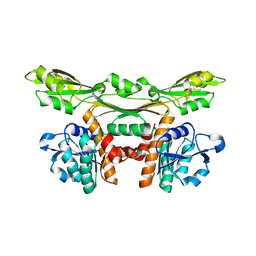 | | Structure of a meso-diaminopimelate dehydrogenase | | Descriptor: | Meso-diaminopimelate D-dehydrogenase | | Authors: | Sakuraba, H, Akita, H, Ohshima, T. | | Deposit date: | 2014-08-25 | | Release date: | 2015-05-06 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Structural insight into the thermostable NADP(+)-dependent meso-diaminopimelate dehydrogenase from Ureibacillus thermosphaericus
Acta Crystallogr.,Sect.D, 71, 2015
|
|
3WYC
 
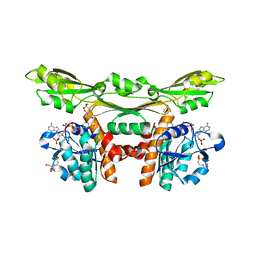 | | Structure of a meso-diaminopimelate dehydrogenase in complex with NADP | | Descriptor: | 2-(2-HYDROXY-1,1-DIHYDROXYMETHYL-ETHYLAMINO)-ETHANESULFONIC ACID, Meso-diaminopimelate D-dehydrogenase, NADP NICOTINAMIDE-ADENINE-DINUCLEOTIDE PHOSPHATE | | Authors: | Sakuraba, H, Akita, H, Ohshima, T. | | Deposit date: | 2014-08-25 | | Release date: | 2015-05-06 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (2.07 Å) | | Cite: | Structural insight into the thermostable NADP(+)-dependent meso-diaminopimelate dehydrogenase from Ureibacillus thermosphaericus
Acta Crystallogr.,Sect.D, 71, 2015
|
|
3VNS
 
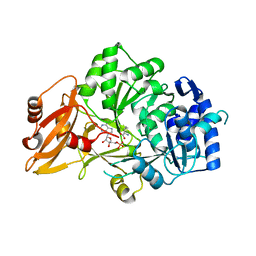 | |
3VNQ
 
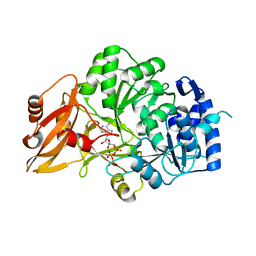 | |
8GQ1
 
 | | HyHEL10 Fab complexed with hen egg lysozyme carrying arginine cluster in framework region of light chain. | | Descriptor: | Heavy Chain of HyHel10 Antibody Fragment (Fab), Light Chain of HyHel10 Antibody Fragment (Fab), Lysozyme C, ... | | Authors: | Matsuura, H, Hirata, K, Sakai, N, Nakakido, M, Tsumoto, K. | | Deposit date: | 2022-08-28 | | Release date: | 2023-08-30 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (3.13 Å) | | Cite: | Arginine cluster introduction on framework region in anti-lysozyme antibody improved association rate constant by changing conformational diversity of CDR loops.
Protein Sci., 32, 2023
|
|
5YAE
 
 | | Ferulic acid esterase from Streptomyces cinnamoneus at 2.4 A resolution | | Descriptor: | ACETATE ION, Esterase, SULFATE ION | | Authors: | Tamura, H, Uraji, M, Mizohata, E, Ogawa, K, Inoue, T, Hatanaka, T. | | Deposit date: | 2017-08-31 | | Release date: | 2017-12-06 | | Last modified: | 2018-01-31 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Loop of Streptomyces Feruloyl Esterase Plays an Important Role in the Enzyme's Catalyzing the Release of Ferulic Acid from Biomass.
Appl. Environ. Microbiol., 84, 2018
|
|
5YAL
 
 | | Ferulic acid esterase from Streptomyces cinnamoneus at 1.5 A resolution | | Descriptor: | ACETATE ION, Esterase, GLYCEROL, ... | | Authors: | Tamura, H, Uraji, M, Mizohata, E, Ogawa, K, Inoue, T, Hatanaka, T. | | Deposit date: | 2017-09-01 | | Release date: | 2017-12-06 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Loop of Streptomyces Feruloyl Esterase Plays an Important Role in the Enzyme's Catalyzing the Release of Ferulic Acid from Biomass.
Appl. Environ. Microbiol., 84, 2018
|
|
3WID
 
 | | Structure of a glucose dehydrogenase T277F mutant in complex with NADP | | Descriptor: | Glucose 1-dehydrogenase, NADP NICOTINAMIDE-ADENINE-DINUCLEOTIDE PHOSPHATE, PENTAETHYLENE GLYCOL, ... | | Authors: | Sakuraba, H, Kanoh, Y, Yoneda, K, Ohshima, T. | | Deposit date: | 2013-09-10 | | Release date: | 2014-05-14 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.25 Å) | | Cite: | Structural insight into glucose dehydrogenase from the thermoacidophilic archaeon Thermoplasma volcanium.
Acta Crystallogr.,Sect.D, 70, 2014
|
|
8I4J
 
 | |
8I4K
 
 | | Structure of Azami Red1.0, a red fluorescent protein engineered from Azami Green | | Descriptor: | Azami Red1.0, CALCIUM ION | | Authors: | Otsubo, S, Takekawa, N, Imamura, H, Imada, K. | | Deposit date: | 2023-01-19 | | Release date: | 2023-11-01 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (1.84 Å) | | Cite: | Red fluorescent proteins engineered from green fluorescent proteins.
Proc.Natl.Acad.Sci.USA, 120, 2023
|
|
3WIC
 
 | | Structure of a substrate/cofactor-unbound glucose dehydrogenase | | Descriptor: | Glucose 1-dehydrogenase, PENTAETHYLENE GLYCOL, S-1,2-PROPANEDIOL, ... | | Authors: | Sakuraba, H, Kanoh, Y, Yoneda, K, Ohshima, T. | | Deposit date: | 2013-09-10 | | Release date: | 2014-05-14 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Structural insight into glucose dehydrogenase from the thermoacidophilic archaeon Thermoplasma volcanium.
Acta Crystallogr.,Sect.D, 70, 2014
|
|
8J1C
 
 | |
8J1G
 
 | | Structure of amino acid dehydrogenase in complex with NADPH | | Descriptor: | 1,2-ETHANEDIOL, ARGININE, NADPH DIHYDRO-NICOTINAMIDE-ADENINE-DINUCLEOTIDE PHOSPHATE, ... | | Authors: | Sakuraba, H, Ohshima, T. | | Deposit date: | 2023-04-12 | | Release date: | 2023-08-16 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | First crystal structure of an NADP + -dependent l-arginine dehydrogenase belonging to the mu-crystallin family.
Int.J.Biol.Macromol., 249, 2023
|
|
3WIE
 
 | | Structure of a glucose dehydrogenase T277F mutant in complex with D-glucose and NAADP | | Descriptor: | Glucose 1-dehydrogenase, ZINC ION, [[(2R,3R,4R,5R)-5-(6-aminopurin-9-yl)-3-oxidanyl-4-phosphonooxy-oxolan-2-yl]methoxy-oxidanyl-phosphoryl] [(2R,3S,4R,5R)-5-(3-carboxypyridin-1-ium-1-yl)-3,4-bis(oxidanyl)oxolan-2-yl]methyl phosphate, ... | | Authors: | Sakuraba, H, Kanoh, Y, Yoneda, K, Ohshima, T. | | Deposit date: | 2013-09-10 | | Release date: | 2014-05-14 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.33 Å) | | Cite: | Structural insight into glucose dehydrogenase from the thermoacidophilic archaeon Thermoplasma volcanium.
Acta Crystallogr.,Sect.D, 70, 2014
|
|
5WYA
 
 | | Structure of amino acid racemase, 2.65 A | | Descriptor: | (2S,3S)-3-methyl-2-[[2-methyl-3-oxidanyl-5-(phosphonooxymethyl)pyridin-4-yl]methylamino]pentanoic acid, DIMETHYL SULFOXIDE, Isoleucine 2-epimerase | | Authors: | Sakuraba, H, Mutaguchi, Y, Hayashi, J, Ohshima, T. | | Deposit date: | 2017-01-11 | | Release date: | 2017-04-19 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (2.65 Å) | | Cite: | Crystal structure of the novel amino-acid racemase isoleucine 2-epimerase from Lactobacillus buchneri.
Acta Crystallogr D Struct Biol, 73, 2017
|
|
5WYF
 
 | | Structure of amino acid racemase, 2.12 A | | Descriptor: | CADMIUM ION, Isoleucine 2-epimerase, N-[O-PHOSPHONO-PYRIDOXYL]-ISOLEUCINE | | Authors: | Sakuraba, H, Mutaguchi, Y, Hayashi, J, Ohshima, T. | | Deposit date: | 2017-01-12 | | Release date: | 2017-04-19 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.12 Å) | | Cite: | Crystal structure of the novel amino-acid racemase isoleucine 2-epimerase from Lactobacillus buchneri.
Acta Crystallogr D Struct Biol, 73, 2017
|
|
2ZA0
 
 | | Crystal structure of mouse glyoxalase I complexed with methyl-gerfelin | | Descriptor: | Glyoxalase I, ZINC ION, methyl 4-(2,3-dihydroxy-5-methylphenoxy)-2-hydroxy-6-methylbenzoate | | Authors: | Okumura, H, Kawatani, M, Osada, H. | | Deposit date: | 2007-09-26 | | Release date: | 2008-08-12 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | The identification of an osteoclastogenesis inhibitor through the inhibition of glyoxalase I
Proc.Natl.Acad.Sci.Usa, 105, 2008
|
|
