8H7Z
 
 | |
8H7L
 
 | | Cryo-EM Structure of SARS-CoV-2 BA.2 Spike protein in complex with BA7535 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, BA7535 fab heavt chain, ... | | Authors: | Liu, Z, Yan, A, Gao, Y. | | Deposit date: | 2022-10-20 | | Release date: | 2023-08-30 | | Last modified: | 2024-03-13 | | Method: | ELECTRON MICROSCOPY (2.44 Å) | | Cite: | Identification of a highly conserved neutralizing epitope within the RBD region of diverse SARS-CoV-2 variants.
Nat Commun, 15, 2024
|
|
8G7B
 
 | |
8G76
 
 | | SARS-CoV-2 spike/Nb5 complex | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Nanosota-5, ... | | Authors: | Ye, G, Bu, F, Liu, B, Li, F. | | Deposit date: | 2023-02-16 | | Release date: | 2024-02-21 | | Method: | ELECTRON MICROSCOPY (3.8 Å) | | Cite: | SARS-CoV-2 spike/Nb5 complex
To Be Published
|
|
8G77
 
 | | SARS-CoV-2 spike/Nb6 complex | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Nanosota-6, ... | | Authors: | Ye, G, Bu, F, Liu, B, Li, F. | | Deposit date: | 2023-02-16 | | Release date: | 2024-02-21 | | Method: | ELECTRON MICROSCOPY (2.8 Å) | | Cite: | SARS-CoV-2 spike/Nb6 complex
To Be Published
|
|
8G7A
 
 | |
8G7C
 
 | |
8G70
 
 | | SARS-CoV-2 spike/nanobody mixture complex | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Nanosota-3, ... | | Authors: | Ye, G, Bu, F, Liu, B, Li, F. | | Deposit date: | 2023-02-16 | | Release date: | 2024-02-21 | | Method: | ELECTRON MICROSCOPY (3.4 Å) | | Cite: | SARS-CoV-2 spike/nanobody mixture complex
To Be Published
|
|
8G78
 
 | | Local refinement of SARS-CoV-2 spike/nanobody mixture complex around NTD | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, Nanosota-5, Nanosota-6, ... | | Authors: | Ye, G, Bu, F, Liu, B, Li, F. | | Deposit date: | 2023-02-16 | | Release date: | 2024-02-21 | | Method: | ELECTRON MICROSCOPY (3.4 Å) | | Cite: | Local refinement of SARS-CoV-2 spike/nanobody mixture complex around NTD
To Be Published
|
|
8G72
 
 | | SARS-CoV-2 spike/Nb2 complex with 1 RBD up (local refinement at 5.6 A) | | Descriptor: | Nanosota-2, Spike glycoprotein | | Authors: | Ye, G, Bu, F, Liu, B, Li, F. | | Deposit date: | 2023-02-16 | | Release date: | 2024-01-24 | | Method: | ELECTRON MICROSCOPY (5.6 Å) | | Cite: | Discovery of Nanosota-2, -3, and -4 as super potent and broad-spectrum therapeutic nanobody candidates against COVID-19.
J.Virol., 97, 2023
|
|
8G74
 
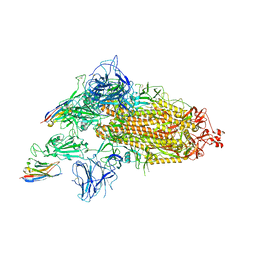 | | SARS-CoV-2 spike/Nb3 complex with 1 RBD up and 2 Nb3 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Nanosota-3, ... | | Authors: | Ye, G, Bu, F, Liu, B, Li, F. | | Deposit date: | 2023-02-16 | | Release date: | 2024-01-24 | | Method: | ELECTRON MICROSCOPY (2.5 Å) | | Cite: | Discovery of Nanosota-2, -3, and -4 as super potent and broad-spectrum therapeutic nanobody candidates against COVID-19.
J.Virol., 97, 2023
|
|
8G73
 
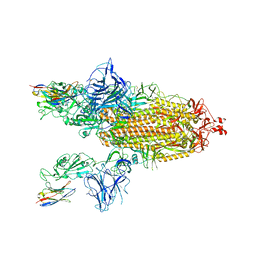 | | SARS-CoV-2 spike/Nb3 complex with 2 RBDs up and 3 Nb3 bound at 2.5 A | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Nanosota-3, ... | | Authors: | Ye, G, Bu, F, Liu, B, Li, F. | | Deposit date: | 2023-02-16 | | Release date: | 2024-01-24 | | Method: | ELECTRON MICROSCOPY (2.5 Å) | | Cite: | Discovery of Nanosota-2, -3, and -4 as super potent and broad-spectrum therapeutic nanobody candidates against COVID-19.
J.Virol., 97, 2023
|
|
8G75
 
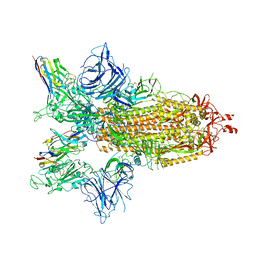 | | SARS-CoV-2 spike/Nb4 complex with 2 RBDs up and 3 Nb4 bound | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Nanosota-4, ... | | Authors: | Ye, G, Bu, F, Liu, B, Li, F. | | Deposit date: | 2023-02-16 | | Release date: | 2024-01-24 | | Method: | ELECTRON MICROSCOPY (3.4 Å) | | Cite: | Discovery of Nanosota-2, -3, and -4 as super potent and broad-spectrum therapeutic nanobody candidates against COVID-19.
J.Virol., 97, 2023
|
|
8G71
 
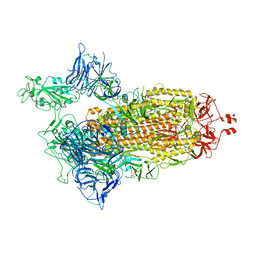 | | Spike/Nb2 complex with 1 RBD up | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Spike glycoprotein | | Authors: | Ye, G, Bu, F, Liu, B, Li, F. | | Deposit date: | 2023-02-16 | | Release date: | 2024-02-21 | | Method: | ELECTRON MICROSCOPY (2.1 Å) | | Cite: | SARS-CoV-2 spike/Nb2 complex with 1 RBD up
To Be Published
|
|
8G79
 
 | |
4ZRT
 
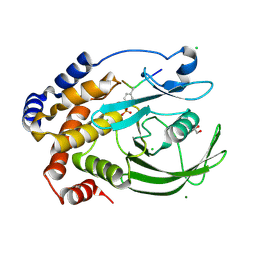 | | PTP1BC215S bound to Nephrin peptide substrate | | Descriptor: | CHLORIDE ION, GLY-PRO-LEU-PTR-ASP-GLU, GLYCEROL, ... | | Authors: | Selner, N.G, Bell, C.E, Pei, D. | | Deposit date: | 2015-05-12 | | Release date: | 2015-06-24 | | Last modified: | 2019-12-25 | | Method: | X-RAY DIFFRACTION (1.74 Å) | | Cite: | Diverse levels of sequence selectivity and catalytic efficiency of protein-tyrosine phosphatases.
Biochemistry, 53, 2014
|
|
4D9L
 
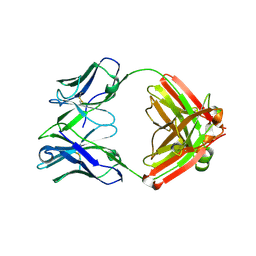 | | Fab structure of anti-HIV-1 gp120 V2 mAb 697 | | Descriptor: | Heavy chain of Fab fragment of anti-HIV1 gp120 V2 mAb 697, Light chain of Fab fragment of anti-HIV1 gp120 V2 mAb 697 | | Authors: | Pan, R.M, Kong, X.P. | | Deposit date: | 2012-01-11 | | Release date: | 2012-03-21 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (2.485 Å) | | Cite: | Functional and immunochemical cross-reactivity of V2-specific monoclonal antibodies from HIV-1-infected individuals.
Virology, 427, 2012
|
|
4F57
 
 | |
4F58
 
 | |
4PV9
 
 | | Crystal Structure of H2Kb-Q600V complex | | Descriptor: | ACETATE ION, Beta-2-microglobulin, GLYCEROL, ... | | Authors: | Twist, K.-A, Rossjohn, J, Gras, S. | | Deposit date: | 2014-03-15 | | Release date: | 2014-04-23 | | Last modified: | 2023-12-06 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Structural and functional correlates of enhanced antiviral immunity generated by heteroclitic CD8 T cell epitopes.
J.Immunol., 192, 2014
|
|
4PG2
 
 | |
4PV8
 
 | | Crystal Structure of H2Kb-Q600F complex | | Descriptor: | ACETATE ION, Beta-2-microglobulin, H-2 class I histocompatibility antigen, ... | | Authors: | Twist, K.-A, Rossjohn, J, Gras, S. | | Deposit date: | 2014-03-15 | | Release date: | 2014-04-23 | | Last modified: | 2023-12-06 | | Method: | X-RAY DIFFRACTION (2.31 Å) | | Cite: | Structural and functional correlates of enhanced antiviral immunity generated by heteroclitic CD8 T cell epitopes.
J.Immunol., 192, 2014
|
|
7RGS
 
 | | The crystal structure of RocC, containing FinO domain, 24-126 | | Descriptor: | Repressor of competence, RNA Chaperone | | Authors: | Kim, H.J, Edwards, R.A, Glover, J.N.M. | | Deposit date: | 2021-07-15 | | Release date: | 2022-11-09 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Structural basis for recognition of transcriptional terminator structures by ProQ/FinO domain RNA chaperones.
Nat Commun, 13, 2022
|
|
7RGT
 
 | | The crystal structure of RocC, containing FinO domain, 1-126 | | Descriptor: | Repressor of competence, RNA Chaperone, SULFATE ION | | Authors: | Kim, H.J, Edwards, R.A, Glover, J.N.M. | | Deposit date: | 2021-07-15 | | Release date: | 2022-11-09 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (2.02 Å) | | Cite: | Structural basis for recognition of transcriptional terminator structures by ProQ/FinO domain RNA chaperones.
Nat Commun, 13, 2022
|
|
7RGU
 
 | |
