3POJ
 
 | | Crystal structure of MASP-1 CUB2 domain bound to Ethylamine | | Descriptor: | 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, CALCIUM ION, ETHANAMINE, ... | | Authors: | Gingras, A.R, Moody, P.C.E, Wallis, R. | | Deposit date: | 2010-11-22 | | Release date: | 2011-08-24 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (1.451 Å) | | Cite: | Structural Basis of Mannan-Binding Lectin Recognition by Its Associated Serine Protease MASP-1: Implications for Complement Activation.
Structure, 19, 2011
|
|
3POF
 
 | | Crystal structure of MASP-1 CUB2 domain bound to Ca2+ | | Descriptor: | 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, CALCIUM ION, Mannan-binding lectin serine protease 1, ... | | Authors: | Gingras, A.R, Moody, P.C.E, Wallis, R. | | Deposit date: | 2010-11-22 | | Release date: | 2011-08-24 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (1.501 Å) | | Cite: | Structural Basis of Mannan-Binding Lectin Recognition by Its Associated Serine Protease MASP-1: Implications for Complement Activation.
Structure, 19, 2011
|
|
3POD
 
 | |
3POI
 
 | | Crystal structure of MASP-1 CUB2 domain bound to Methylamine | | Descriptor: | 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, CALCIUM ION, METHYLAMINE, ... | | Authors: | Gingras, A.R, Moody, P.C.E, Wallis, R. | | Deposit date: | 2010-11-22 | | Release date: | 2011-08-24 | | Last modified: | 2011-11-30 | | Method: | X-RAY DIFFRACTION (1.701 Å) | | Cite: | Structural Basis of Mannan-Binding Lectin Recognition by Its Associated Serine Protease MASP-1: Implications for Complement Activation.
Structure, 19, 2011
|
|
3POB
 
 | |
3POG
 
 | |
3PON
 
 | |
3POE
 
 | |
1LA6
 
 | | The crystal structure of Trematomus newnesi hemoglobin in a partial hemichrome state | | Descriptor: | CARBON MONOXIDE, Hemoglobin alpha-1 chain, Hemoglobin beta-1/2 chain, ... | | Authors: | Riccio, A, Vitagliano, L, di Prisco, G, Zagari, A, Mazzarella, L. | | Deposit date: | 2002-03-28 | | Release date: | 2002-07-31 | | Last modified: | 2023-08-16 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | The crystal structure of a tetrameric hemoglobin in a partial hemichrome state
Proc.Natl.Acad.Sci.USA, 99, 2002
|
|
1T1N
 
 | | CRYSTAL STRUCTURE OF CARBONMONOXY HEMOGLOBIN | | Descriptor: | CARBON MONOXIDE, PROTEIN (HEMOGLOBIN), PROTOPORPHYRIN IX CONTAINING FE | | Authors: | Mazzarella, L, Vitagliano, L, Savino, C, Zagari, A. | | Deposit date: | 1999-03-05 | | Release date: | 1999-04-29 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Crystal structure of Trematomus newnesi haemoglobin re-opens the root effect question.
J.Mol.Biol., 287, 1999
|
|
2AA1
 
 | | Crystal structure of the cathodic hemoglobin isolated from the Antarctic fish Trematomus Newnesi | | Descriptor: | Hemoglobin alpha-1 chain, Hemoglobin beta-C chain, PROTOPORPHYRIN IX CONTAINING FE | | Authors: | Mazzarella, L, Bonomi, G, Lubrano, M.C, Merlino, A, Riccio, A, Vergara, A, Vitagliano, L, Verde, C, Di Prisco, G. | | Deposit date: | 2005-07-13 | | Release date: | 2005-08-02 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Minimal structural requirements for root effect: crystal structure of the cathodic hemoglobin isolated from the antarctic fish Trematomus newnesi
Proteins, 62, 2006
|
|
2X08
 
 | | cytochrome c peroxidase: ascorbate bound to the engineered ascorbate binding site | | Descriptor: | ASCORBIC ACID, CYTOCHROME C PEROXIDASE, MITOCHONDRIAL, ... | | Authors: | Murphy, E.J, Metcalfe, C.L, Gumiero, A, Raven, E.L, Moody, P.C.E. | | Deposit date: | 2009-12-07 | | Release date: | 2010-11-10 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2.01 Å) | | Cite: | Engineering the substrate specificity and reactivity of a heme protein: creation of an ascorbate binding site in cytochrome c peroxidase.
Biochemistry, 47, 2008
|
|
2XIF
 
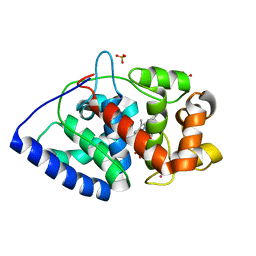 | | The structure of ascorbate peroxidase Compound II | | Descriptor: | ASCORBATE PEROXIDASE, POTASSIUM ION, PROTOPORPHYRIN IX CONTAINING FE, ... | | Authors: | Gumiero, A, Raven, E.L, Moody, P.C.E. | | Deposit date: | 2010-06-29 | | Release date: | 2010-07-07 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (1.65 Å) | | Cite: | Nature of the ferryl heme in compounds I and II.
J. Biol. Chem., 286, 2011
|
|
2X07
 
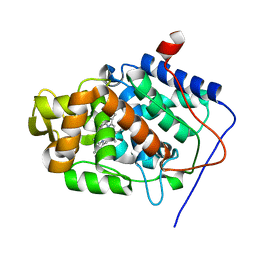 | | cytochrome c peroxidase: engineered ascorbate binding site | | Descriptor: | Cytochrome c peroxidase, mitochondrial, PROTOPORPHYRIN IX CONTAINING FE | | Authors: | Murphy, E.J, Metcalfe, C.L, Gumiero, A, Raven, E.L, Moody, P.C.E. | | Deposit date: | 2009-12-07 | | Release date: | 2010-11-10 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (1.86 Å) | | Cite: | Engineering the substrate specificity and reactivity of a heme protein: creation of an ascorbate binding site in cytochrome c peroxidase.
Biochemistry, 47, 2008
|
|
2XIL
 
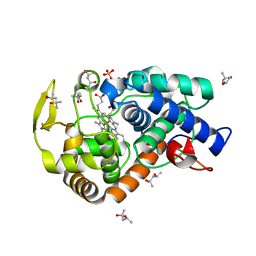 | | The structure of cytochrome c peroxidase Compound I | | Descriptor: | (4R)-2-METHYLPENTANE-2,4-DIOL, (4S)-2-METHYL-2,4-PENTANEDIOL, CYTOCHROME C PEROXIDASE, ... | | Authors: | Gumiero, A, Raven, E.L, Moody, P.C.E. | | Deposit date: | 2010-06-30 | | Release date: | 2010-07-14 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (1.68 Å) | | Cite: | Nature of the ferryl heme in compounds I and II.
J. Biol. Chem., 286, 2011
|
|
2XJ8
 
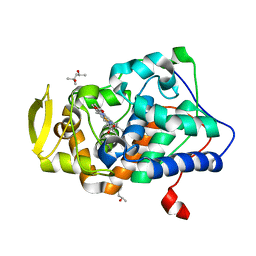 | | The structure of ferrous cytochrome c peroxidase | | Descriptor: | (4S)-2-METHYL-2,4-PENTANEDIOL, CYTOCHROME C PEROXIDASE, MITOCHONDRIAL, ... | | Authors: | Gumiero, A, Raven, E.L, Moody, P.C.E. | | Deposit date: | 2010-07-02 | | Release date: | 2010-07-14 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (1.69 Å) | | Cite: | Nature of the ferryl heme in compounds I and II.
J. Biol. Chem., 286, 2011
|
|
2XJ5
 
 | | The structure of cytochrome c peroxidase Compound II | | Descriptor: | (4S)-2-METHYL-2,4-PENTANEDIOL, CYTOCHROME C PEROXIDASE, MITOCHONDRIAL, ... | | Authors: | Gumiero, A, Raven, E.L, Moody, P.C.E. | | Deposit date: | 2010-07-02 | | Release date: | 2010-07-14 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (1.69 Å) | | Cite: | Nature of the ferryl heme in compounds I and II.
J. Biol. Chem., 286, 2011
|
|
2XI6
 
 | | The structure of ascorbate peroxidase Compound I | | Descriptor: | ASCORBATE PEROXIDASE, POTASSIUM ION, PROTOPORPHYRIN IX CONTAINING FE, ... | | Authors: | Gumiero, A, Raven, E.L, Moody, P.C.E. | | Deposit date: | 2010-06-29 | | Release date: | 2010-07-14 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (1.65 Å) | | Cite: | Nature of the ferryl heme in compounds I and II.
J. Biol. Chem., 286, 2011
|
|
2XIH
 
 | | The structure of ascorbate peroxidase Compound III | | Descriptor: | ASCORBATE PEROXIDASE, OXYGEN MOLECULE, POTASSIUM ION, ... | | Authors: | Gumiero, A, Raven, E.L, Moody, P.C.E. | | Deposit date: | 2010-06-29 | | Release date: | 2010-07-07 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (1.65 Å) | | Cite: | Nature of the ferryl heme in compounds I and II.
J. Biol. Chem., 286, 2011
|
|
2XJ6
 
 | | The structure of ferrous ascorbate peroxidase | | Descriptor: | ASCORBATE PEROXIDASE, POTASSIUM ION, PROTOPORPHYRIN IX CONTAINING FE, ... | | Authors: | Gumiero, A, Raven, E.L, Moody, P.C.E. | | Deposit date: | 2010-07-02 | | Release date: | 2010-07-14 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Nature of the ferryl heme in compounds I and II.
J. Biol. Chem., 286, 2011
|
|
1H63
 
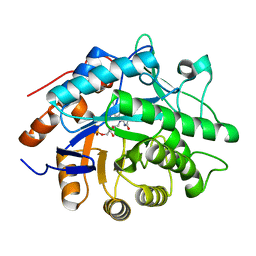 | | Structure of the reduced Pentaerythritol Tetranitrate Reductase | | Descriptor: | FLAVIN MONONUCLEOTIDE, PENTAERYTHRITOL TETRANITRATE REDUCTASE | | Authors: | Barna, T.M, Moody, P.C.E. | | Deposit date: | 2001-06-04 | | Release date: | 2001-07-05 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (1.62 Å) | | Cite: | Crystal Structure of Pentaerythritol Tetranitrate Reductase: "Flipped" Binding Geometries for Steroid Substrates in Different Redox States of the Enzyme
J.Mol.Biol., 310, 2001
|
|
1H60
 
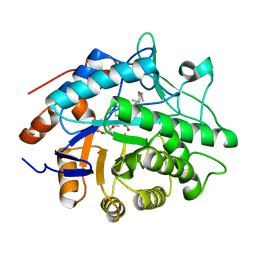 | |
1H62
 
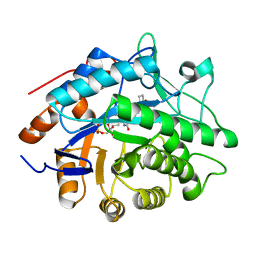 | | Structure of Pentaerythritol tetranitrate reductase in complex with 1,4-androstadien-3,17-dione | | Descriptor: | ANDROSTA-1,4-DIENE-3,17-DIONE, FLAVIN MONONUCLEOTIDE, PENTAERYTHRITOL TETRANITRATE REDUCTASE | | Authors: | Barna, T.M, Moody, P.C.E. | | Deposit date: | 2001-06-04 | | Release date: | 2001-07-05 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Crystal Structure of Pentaerythritol Tetranitrate Reductase: "Flipped" Binding Geometries for Steroid Substrates in Different Redox States of the Enzyme
J.Mol.Biol., 310, 2001
|
|
1H61
 
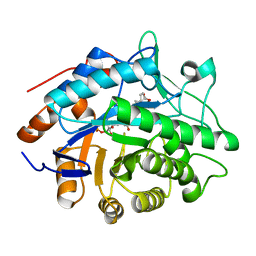 | | Structure of Pentaerythritol Tetranitrate Reductase in complex with prednisone | | Descriptor: | 17,21-DIHYDROXYPREGNA-1,4-DIENE-3,11,20-TRIONE, FLAVIN MONONUCLEOTIDE, PENTAERYTHRITOL TETRANITRATE REDUCTASE | | Authors: | Barna, T.M, Moody, P.C.E. | | Deposit date: | 2001-06-04 | | Release date: | 2001-07-05 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (1.4 Å) | | Cite: | Crystal Structure of Pentaerythritol Tetranitrate Reductase: "Flipped" Binding Geometries for Steroid Substrates in Different Redox States of the Enzyme
J.Mol.Biol., 310, 2001
|
|
1GD1
 
 | |
