7SQO
 
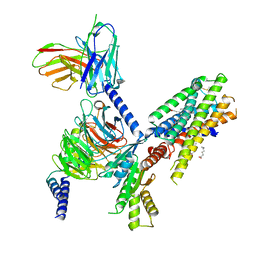 | | Structure of the orexin-2 receptor(OX2R) bound to TAK-925, Gi and scFv16 | | Descriptor: | Guanine nucleotide-binding protein G(I)/G(S)/G(O) subunit gamma-2, Guanine nucleotide-binding protein G(I)/G(S)/G(T) subunit beta-1, Guanine nucleotide-binding protein G(i) subunit alpha-1, ... | | Authors: | McGrath, A.P, Kang, Y, Flinspach, M. | | Deposit date: | 2021-11-05 | | Release date: | 2022-05-25 | | Last modified: | 2022-07-06 | | Method: | ELECTRON MICROSCOPY (3.17 Å) | | Cite: | Molecular mechanism of the wake-promoting agent TAK-925.
Nat Commun, 13, 2022
|
|
7SR8
 
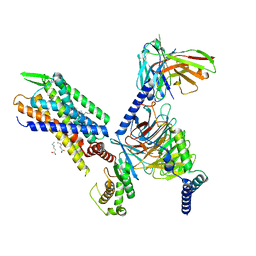 | | Molecular mechanism of the the wake-promoting agent TAK-925 | | Descriptor: | Guanine nucleotide-binding protein G(I)/G(S)/G(O) subunit gamma-2, Guanine nucleotide-binding protein G(I)/G(S)/G(T) subunit beta-1, Hypocretin receptor type 2, ... | | Authors: | Yin, J, Chapman, K, Lian, P, De Brabander, J.K, Rosenbaum, D.M. | | Deposit date: | 2021-11-08 | | Release date: | 2022-06-08 | | Method: | ELECTRON MICROSCOPY (3.3 Å) | | Cite: | Molecular mechanism of the wake-promoting agent TAK-925.
Nat Commun, 13, 2022
|
|
3VT4
 
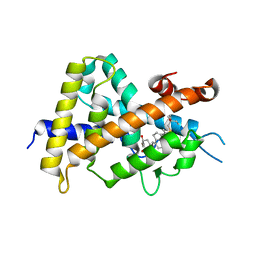 | | Crystal structures of rat VDR-LBD with R270L mutation | | Descriptor: | (1R,2Z,3R,5E,7E)-17-{(1S)-1-[(2-ethyl-2-hydroxybutyl)sulfanyl]ethyl}-2-(2-hydroxyethylidene)-9,10-secoestra-5,7,16-triene-1,3-diol, COACTIVATOR PEPTIDE DRIP, Vitamin D3 receptor | | Authors: | Nakabayashi, M, Shimizu, M, Ikura, T, Ito, N. | | Deposit date: | 2012-05-19 | | Release date: | 2013-05-22 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Crystal structures of hereditary vitamin D-resistant rickets-associated vitamin D receptor mutants R270L and W282R bound to 1,25-dihydroxyvitamin D3 and synthetic ligands.
J.Med.Chem., 56, 2013
|
|
3VT7
 
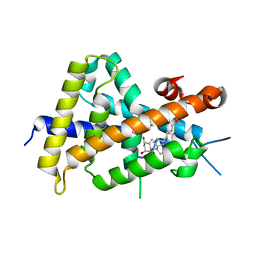 | | Crystal structures of rat VDR-LBD with W282R mutation | | Descriptor: | 5-{2-[1-(5-HYDROXY-1,5-DIMETHYL-HEXYL)-7A-METHYL-OCTAHYDRO-INDEN-4-YLIDENE]-ETHYLIDENE}-4-METHYLENE-CYCLOHEXANE-1,3-DIOL, COACTIVATOR PEPTIDE DRIP, Vitamin D3 receptor | | Authors: | Nakabayashi, M, Shimizu, M, Ikura, T, Ito, N. | | Deposit date: | 2012-05-19 | | Release date: | 2013-05-22 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (1.65 Å) | | Cite: | Crystal structures of hereditary vitamin D-resistant rickets-associated vitamin D receptor mutants R270L and W282R bound to 1,25-dihydroxyvitamin D3 and synthetic ligands.
J.Med.Chem., 56, 2013
|
|
2RUF
 
 | |
2RUE
 
 | |
3TZ1
 
 | | Crystal structure of the Ca2+-saturated C-terminal domain of Akazara scallop troponin C in complex with a troponin I fragment | | Descriptor: | CALCIUM ION, Troponin C, Troponin I | | Authors: | Yumoto, F, Kato, Y.S, Ohtsuki, I, Tanokura, M. | | Deposit date: | 2011-09-26 | | Release date: | 2013-01-23 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Structure of the Ca2+-saturated C-terminal domain of scallop troponin C in complex with a troponin I fragment
Biol.Chem., 394, 2012
|
|
2EJX
 
 | | Crystal structure of the hypothetical protein STK_08120 from Sulfolobus tokodaii | | Descriptor: | STK_08120 | | Authors: | Miyakawa, T, Miyazono, K, Sawano, Y, Hatano, K, Nagata, K, Tanokura, M. | | Deposit date: | 2007-03-21 | | Release date: | 2008-03-25 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (1.79 Å) | | Cite: | A thermoacidophile-specific protein family, DUF3211, functions as a fatty acid carrier with novel binding mode
J.Bacteriol., 195, 2013
|
|
1WTB
 
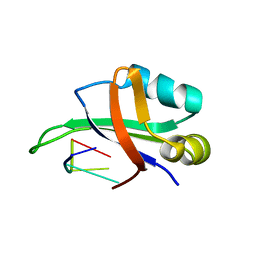 | | Complex structure of the C-terminal RNA-binding domain of hnRNP D (AUF1) with telomere DNA | | Descriptor: | 5'-D(P*TP*AP*GP*G)-3', Heterogeneous nuclear ribonucleoprotein D0 | | Authors: | Enokizono, Y, Konishi, Y, Nagata, K, Ouhashi, K, Uesugi, S, Ishikawa, F, Katahira, M. | | Deposit date: | 2004-11-22 | | Release date: | 2005-04-05 | | Last modified: | 2024-05-29 | | Method: | SOLUTION NMR | | Cite: | Structure of hnRNP D complexed with single-stranded telomere DNA and unfolding of the quadruplex by heterogeneous nuclear ribonucleoprotein D
J.Biol.Chem., 280, 2005
|
|
1X0F
 
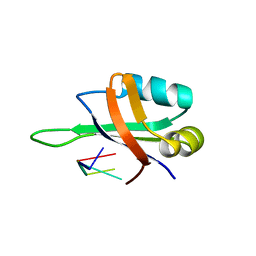 | | Complex structure of the C-terminal RNA-binding domain of hnRNP D(AUF1) with telomeric DNA | | Descriptor: | 5'-D(P*TP*AP*GP*G)-3', Heterogeneous nuclear ribonucleoprotein D0 | | Authors: | Enokizono, Y, Konishi, Y, Nagata, K, Ouhashi, K, Uesugi, S, Ishikawa, F, Katahira, M. | | Deposit date: | 2005-03-22 | | Release date: | 2005-04-05 | | Last modified: | 2024-05-29 | | Method: | SOLUTION NMR | | Cite: | Structure of hnRNP D complexed with single-stranded telomere DNA and unfolding of the quadruplex by heterogeneous nuclear ribonucleoprotein D.
J.Biol.Chem., 280, 2005
|
|
3OPM
 
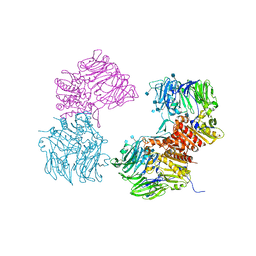 | | Crystal Structure of Human DPP4 Bound to TAK-294 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, 2-{[3-(aminomethyl)-2-(2-methylpropyl)-1-oxo-4-phenyl-1,2-dihydroisoquinolin-6-yl]oxy}acetamide, ... | | Authors: | Yano, J.K, Aertgeerts, K. | | Deposit date: | 2010-09-01 | | Release date: | 2011-10-19 | | Last modified: | 2020-07-29 | | Method: | X-RAY DIFFRACTION (2.72 Å) | | Cite: | Identification of 3-aminomethyl-1,2-dihydro-4-phenyl-1-isoquinolones: a new class of potent, selective, and orally active non-peptide dipeptidyl peptidase IV inhibitors that form a unique interaction with Lys554.
Bioorg.Med.Chem., 19, 2011
|
|
2JMW
 
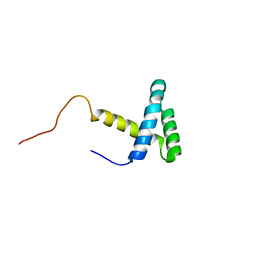 | | Structure of DNA-Binding Domain of Arabidopsis GT-1 | | Descriptor: | DNA binding protein GT-1 | | Authors: | Nagata, T, Niyada, E, Noto, K, Ikeda, Y, Yamamoto, Y, Uesugi, S, Murata, J, Hiratsuka, K, Katahira, M. | | Deposit date: | 2006-12-11 | | Release date: | 2007-12-11 | | Last modified: | 2023-12-20 | | Method: | SOLUTION NMR | | Cite: | Solution structures of the trihelix DNA-binding domains of the wild-type and a phosphomimetic mutant of Arabidopsis GT-1: mechanism for an increase in DNA-binding affinity through phosphorylation.
Proteins, 78, 2010
|
|
3T1F
 
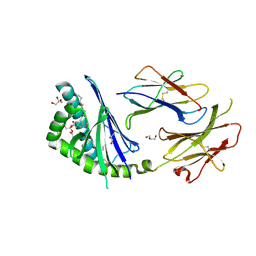 | | Crystal structure of the mouse CD1d-Glc-DAG-s2 complex | | Descriptor: | (2S)-1-(alpha-D-glucopyranosyloxy)-3-(hexadecanoyloxy)propan-2-yl (11Z)-octadec-11-enoate, 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Girardi, E, Zajonc, D.M. | | Deposit date: | 2011-07-21 | | Release date: | 2011-09-21 | | Last modified: | 2020-07-29 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Invariant natural killer T cells recognize glycolipids from pathogenic Gram-positive bacteria.
Nat.Immunol., 12, 2011
|
|
2KTD
 
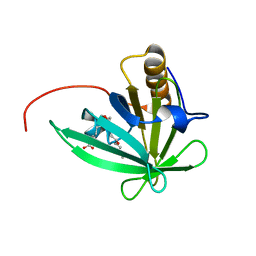 | | Solution structure of mouse lipocalin-type prostaglandin D synthase / substrate analog (U-46619) complex | | Descriptor: | (5Z)-7-{(1R,4S,5S,6R)-6-[(1E,3S)-3-hydroxyoct-1-en-1-yl]-2-oxabicyclo[2.2.1]hept-5-yl}hept-5-enoic acid, Prostaglandin-H2 D-isomerase | | Authors: | Shimamoto, S, Maruo, H, Yoshida, T, Kato, N, Ohkubo, T. | | Deposit date: | 2010-01-27 | | Release date: | 2011-02-02 | | Last modified: | 2024-05-01 | | Method: | SOLUTION NMR | | Cite: | Solution Structure of Lipocalin-type Prostaglandin D synthase / Substrate analog complex reveals Open-Closed Conformational Change required for Substrate Recognition
To be Published
|
|
4YD9
 
 | | Crystal structure of squid hemocyanin | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, CU2-O2 CLUSTER, alpha-D-mannopyranose-(1-3)-[alpha-D-mannopyranose-(1-6)]beta-D-mannopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Matsuno, A, Gai, Z, Kato, K, Tanaka, Y, Yao, M. | | Deposit date: | 2015-02-21 | | Release date: | 2015-10-14 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | Crystal Structure of the 3.8-MDa Respiratory Supermolecule Hemocyanin at 3.0 angstrom Resolution
Structure, 23, 2015
|
|
3H8D
 
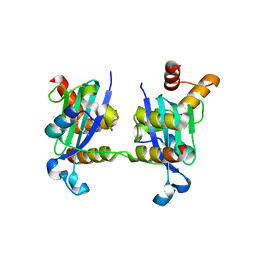 | | Crystal structure of Myosin VI in complex with Dab2 peptide | | Descriptor: | 2,3-DIHYDROXY-1,4-DITHIOBUTANE, CHLORIDE ION, Disabled homolog 2, ... | | Authors: | Yu, C, Feng, W, Wei, Z, Zhang, M. | | Deposit date: | 2009-04-29 | | Release date: | 2009-09-29 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Myosin VI undergoes cargo-mediated dimerization
Cell(Cambridge,Mass.), 138, 2009
|
|
3VVI
 
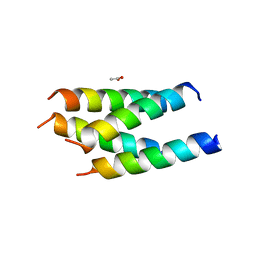 | |
8K9O
 
 | |
2KP1
 
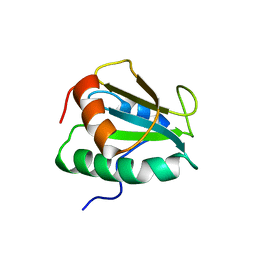 | |
2KP2
 
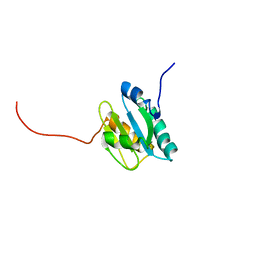 | |
6LCS
 
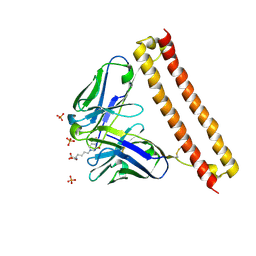 | | Crystal structure of 73MuL9 Fv-clasp fragment in complex with GA-pyridine analogue | | Descriptor: | (2~{S})-6-[4-(hydroxymethyl)-3-oxidanyl-pyridin-1-ium-1-yl]-2-(phenylmethoxycarbonylamino)hexanoic acid, PHOSPHATE ION, VH-SARAH, ... | | Authors: | Nakamura, T, Takagi, J, Yamagata, Y, Morioka, H. | | Deposit date: | 2019-11-19 | | Release date: | 2020-11-25 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Molecular recognition of a single-chain Fv antibody specific for GA-pyridine, an advanced glycation end-product (AGE), elucidated using biophysical techniques and synthetic antigen analogues.
J.Biochem., 170, 2021
|
|
1WMR
 
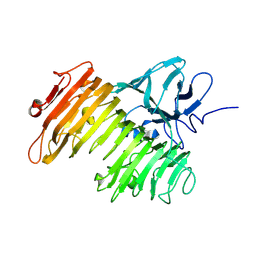 | | Crystal Structure of Isopullulanase from Aspergillus niger ATCC 9642 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, Isopullulanase | | Authors: | Mizuno, M, Tonozuka, T, Miyasaka, Y, Akeboshi, H, Kamitori, S, Nishikawa, A, Sakano, Y. | | Deposit date: | 2004-07-15 | | Release date: | 2005-07-19 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Crystal Structure of Aspergillus niger Isopullulanase, a Member of Glycoside Hydrolase Family 49
J.Mol.Biol., 376, 2008
|
|
1X0C
 
 | | Improved Crystal Structure of Isopullulanase from Aspergillus niger ATCC 9642 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, Isopullulanase | | Authors: | Mizuno, M, Tonozuka, T, Yamamura, A, Miyasaka, Y, Akeboshi, H, Kamitori, S, Nishikawa, A, Sakano, Y. | | Deposit date: | 2005-03-17 | | Release date: | 2006-06-13 | | Last modified: | 2020-07-29 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Crystal Structure of Aspergillus niger Isopullulanase, a Member of Glycoside Hydrolase Family 49
J.Mol.Biol., 376, 2008
|
|
5XA4
 
 | | Crystal Structure of HasAp with Fe-5,15-Diazaporphyrin | | Descriptor: | 10,20-Diphenyl-5,15-diaza-porphyrin containing FE, Heme acquisition protein HasAp | | Authors: | Shoji, O, Uehara, H, Sugimoto, H, Shiro, Y, Watanabe, Y. | | Deposit date: | 2017-03-10 | | Release date: | 2017-12-06 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (1.3 Å) | | Cite: | Structures of the Heme Acquisition Protein HasA with Iron(III)-5,15-Diphenylporphyrin and Derivatives Thereof as an Artificial Prosthetic Group
Angew. Chem. Int. Ed. Engl., 56, 2017
|
|
5XIC
 
 | | Crystal Structure of HasAp with Fe-5,10,15-triphenylporphyrin | | Descriptor: | 3,6,9,12,15,18,21-HEPTAOXATRICOSANE-1,23-DIOL, 5,10,15-Triphenylporphyrin cpntaining FE, Heme acquisition protein HasAp | | Authors: | Shoji, O, Uehara, H, Sugimoto, H, Shiro, Y, Watanabe, Y. | | Deposit date: | 2017-04-26 | | Release date: | 2017-12-06 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (1.45 Å) | | Cite: | Structures of the Heme Acquisition Protein HasA with Iron(III)-5,15-Diphenylporphyrin and Derivatives Thereof as an Artificial Prosthetic Group
Angew. Chem. Int. Ed. Engl., 56, 2017
|
|
