1H70
 
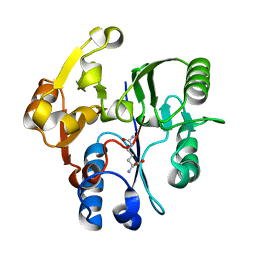 | | DDAH FROM PSEUDOMONAS AERUGINOSA. C249S MUTANT COMPLEXED WITH CITRULLINE | | Descriptor: | CITRULLINE, NG, NG-DIMETHYLARGININE DIMETHYLAMINOHYDROLASE | | Authors: | Murray-Rust, J, Leiper, J, McAlister, M, Phelan, J, Tilley, S, Santamaria, J, Vallance, P, McDonald, N. | | Deposit date: | 2001-06-30 | | Release date: | 2001-08-02 | | Last modified: | 2024-05-01 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Structural insights into the hydrolysis of cellular nitric oxide synthase inhibitors by dimethylarginine dimethylaminohydrolase.
Nat. Struct. Biol., 8, 2001
|
|
3L45
 
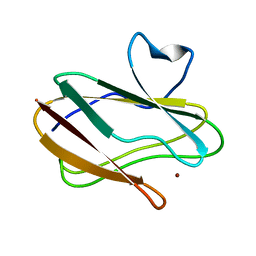 | | A Joint Neutron and X-ray structure of Oxidized Amicyanin | | Descriptor: | Amicyanin, COPPER (II) ION | | Authors: | Sukumar, N, Mathews, F.S, Langan, P, Davidson, V.L. | | Deposit date: | 2009-12-18 | | Release date: | 2010-04-28 | | Last modified: | 2023-09-13 | | Method: | NEUTRON DIFFRACTION (1.8 Å), X-RAY DIFFRACTION | | Cite: | A joint x-ray and neutron study on amicyanin reveals the role of protein dynamics in electron transfer.
Proc.Natl.Acad.Sci.USA, 107, 2010
|
|
4XQD
 
 | | X-ray structure analysis of xylanase-WT at pH4.0 | | Descriptor: | 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, Endo-1,4-beta-xylanase 2, IODIDE ION | | Authors: | Wan, Q, Park, J.M, Riccardi, D.M, Hanson, L.B, Fisher, Z, Smith, J.C, Ostermann, A, Schrader, T, Graham, D.E, Coates, L, Langan, P, Kovalevsky, A.Y. | | Deposit date: | 2015-01-19 | | Release date: | 2015-09-23 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Direct determination of protonation states and visualization of hydrogen bonding in a glycoside hydrolase with neutron crystallography.
Proc.Natl.Acad.Sci.USA, 112, 2015
|
|
4XQW
 
 | | X-ray structure analysis of xylanase-N44E with MES at pH6.0 | | Descriptor: | 2-(N-MORPHOLINO)-ETHANESULFONIC ACID, Endo-1,4-beta-xylanase 2, IODIDE ION | | Authors: | Wan, Q, Park, J.M, Riccardi, D.M, Hanson, L.B, Fisher, Z, Smith, J.C, Ostermann, A, Schrader, T, Graham, D.E, Coates, L, Langan, P, Kovalevsky, A.Y. | | Deposit date: | 2015-01-20 | | Release date: | 2015-09-23 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Direct determination of protonation states and visualization of hydrogen bonding in a glycoside hydrolase with neutron crystallography.
Proc.Natl.Acad.Sci.USA, 112, 2015
|
|
4XPV
 
 | | Neutron and X-ray structure analysis of xylanase: N44D at pH6 | | Descriptor: | Endo-1,4-beta-xylanase 2, IODIDE ION | | Authors: | Wan, Q, Park, J.M, Riccardi, D.M, Hanson, L.B, Fisher, Z, Smith, J.C, Ostermann, A, Schrader, T, Graham, D.E, Coates, L, Langan, P, Kovalevsky, A.Y. | | Deposit date: | 2015-01-18 | | Release date: | 2015-09-30 | | Last modified: | 2023-09-27 | | Method: | NEUTRON DIFFRACTION (1.7 Å), X-RAY DIFFRACTION | | Cite: | Direct determination of protonation states and visualization of hydrogen bonding in a glycoside hydrolase with neutron crystallography.
Proc.Natl.Acad.Sci.USA, 112, 2015
|
|
4XQ4
 
 | | X-ray structure analysis of xylanase - N44D | | Descriptor: | Endo-1,4-beta-xylanase 2, IODIDE ION | | Authors: | Wan, Q, Park, J.M, Riccardi, D.M, Hanson, L.B, Fisher, Z, Smith, J.C, Ostermann, A, Schrader, T, Graham, D.E, Coates, L, Langan, P, Kovalevsky, A.Y. | | Deposit date: | 2015-01-19 | | Release date: | 2015-09-23 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (1.25 Å) | | Cite: | Direct determination of protonation states and visualization of hydrogen bonding in a glycoside hydrolase with neutron crystallography.
Proc.Natl.Acad.Sci.USA, 112, 2015
|
|
6HB4
 
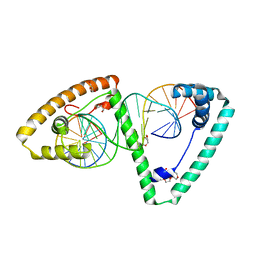 | | TFAM in Complex with Site-Y | | Descriptor: | DNA (5'*CP*TP*GP*TP*GP*CP*AP*GP*AP*CP*AP*TP*TP*CP*AP*AP*TP*TP*GP*TP*TP*A)-3'), DNA (5'-D(*TP*AP*AP*CP*AP*AP*TP*TP*GP*AP*AP*TP*GP*TP*CP*TP*GP*CP*AP*CP*AP*G)-3'), GLYCEROL, ... | | Authors: | Cuppari, A, Fernandez-Millan, P, Rubio-Cosials, A, Tarres-Sole, A, Lyonnais, S, Sola, M. | | Deposit date: | 2018-08-09 | | Release date: | 2019-06-05 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (3.05 Å) | | Cite: | DNA specificities modulate the binding of human transcription factor A to mitochondrial DNA control region.
Nucleic Acids Res., 47, 2019
|
|
3N6S
 
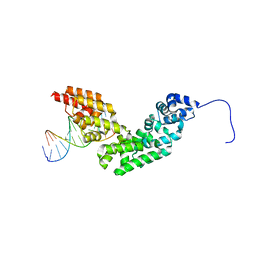 | | Crystal structure of human mitochondrial mTERF in complex with a 15-mer DNA encompassing the tRNALeu(UUR) binding sequence | | Descriptor: | DNA (5'-D(*AP*TP*GP*GP*CP*AP*GP*AP*GP*CP*CP*CP*GP*GP*T)-3'), DNA (5'-D(*TP*AP*CP*CP*GP*GP*GP*CP*TP*CP*TP*GP*CP*CP*A)-3'), Transcription termination factor, ... | | Authors: | Jimenez-Menendez, N, Fernandez-Millan, P, Rubio-Cosials, A, Arnan, C, Montoya, J, Jacobs, H.T, Bernado, P, Coll, M, Uson, I, Sola, M. | | Deposit date: | 2010-05-26 | | Release date: | 2010-06-16 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (3.1 Å) | | Cite: | Human mitochondrial mTERF wraps around DNA through a left-handed superhelical tandem repeat.
Nat.Struct.Mol.Biol., 17, 2010
|
|
7QAA
 
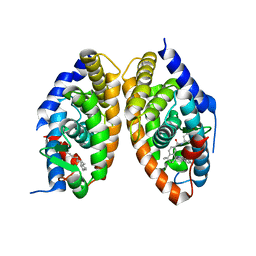 | | Crystal structure of RARalpha/RXRalpha ligand binding domain heterodimer in complex with BMS614 and oleic acid | | Descriptor: | 4-[(4,4-DIMETHYL-1,2,3,4-TETRAHYDRO-[1,2']BINAPTHALENYL-7-CARBONYL)-AMINO]-BENZOIC ACID, Isoform Alpha-1-deltaBC of Retinoic acid receptor alpha, OLEIC ACID, ... | | Authors: | le Maire, A, Vivat, V, Guee, L, Blanc, P, Malosse, C, Chamot-Rooke, J, Germain, P, Bourguet, w. | | Deposit date: | 2021-11-16 | | Release date: | 2022-10-05 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (2.76 Å) | | Cite: | Design and in vitro characterization of RXR variants as tools to investigate the biological role of endogenous rexinoids.
J.Mol.Endocrinol., 69, 2022
|
|
7OD9
 
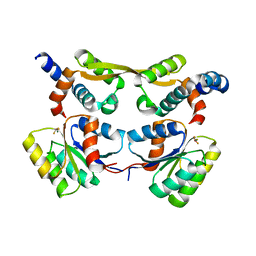 | | Crystal structure of activated CheY fused to the C-terminal domain of CheF | | Descriptor: | BERYLLIUM TRIFLUORIDE ION, C-terminal domain of CheF from Methanococcus maripaludis, MAGNESIUM ION, ... | | Authors: | Altegoer, F, Weiland, P, Bange, G. | | Deposit date: | 2021-04-29 | | Release date: | 2022-04-27 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Structural insights into the mechanism of archaellar rotational switching.
Nat Commun, 13, 2022
|
|
6YMT
 
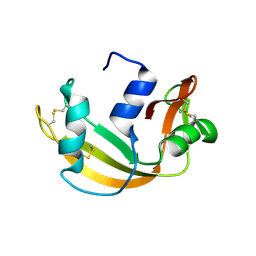 | | RNASE 3/1 version1 | | Descriptor: | GLYCEROL, RNase 3/1 version 1 | | Authors: | Fernandez-Millan, P, Prats-Ejarque, G, Vazquez-Monteagudo, S, Boix, E. | | Deposit date: | 2020-04-09 | | Release date: | 2021-07-21 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (1.58 Å) | | Cite: | Exploring the RNase A scaffold to combine catalytic and antimicrobial activities. Structural characterization of RNase 3/1 chimeras.
Front Mol Biosci, 9, 2022
|
|
6YBC
 
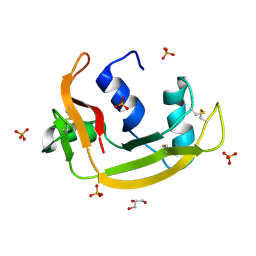 | | RNASE 3/1 version2 phosphate complex | | Descriptor: | GLYCEROL, PHOSPHATE ION, RNASE 3/1 version2 | | Authors: | Fernandez-Millan, P, Prats-Ejarque, G, Vazquez-Monteagudo, S, Boix, E. | | Deposit date: | 2020-03-16 | | Release date: | 2021-10-06 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (1.49 Å) | | Cite: | Exploring the RNase A scaffold to combine catalytic and antimicrobial activities. Structural characterization of RNase 3/1 chimeras.
Front Mol Biosci, 9, 2022
|
|
6YBE
 
 | |
7OVP
 
 | |
3N7Q
 
 | | Crystal structure of human mitochondrial mTERF fragment (aa 99-399) in complex with a 12-mer DNA encompassing the tRNALeu(UUR) binding sequence | | Descriptor: | CITRIC ACID, DNA (5'-D(*CP*CP*GP*GP*GP*CP*TP*CP*TP*GP*CP*C)-3'), DNA (5'-D(*GP*GP*CP*AP*GP*AP*GP*CP*CP*CP*GP*G)-3'), ... | | Authors: | Jimenez-Menendez, N, Fernandez-Millan, P, Rubio-Cosials, A, Arnan, C, Montoya, J, Jacobs, H.T, Bernado, P, Coll, M, Uson, I, Sola, M. | | Deposit date: | 2010-05-27 | | Release date: | 2010-06-16 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Human mitochondrial mTERF wraps around DNA through a left-handed superhelical tandem repeat.
Nat.Struct.Mol.Biol., 17, 2010
|
|
8F11
 
 | | T4 lysozyme with a 2,6-diazaadamantane nitroxide (DZD) spin label | | Descriptor: | 1-[(1r,3r,5r,7r)-6-hydroxy-2,6-diazatricyclo[3.3.1.1~3,7~]decan-2-yl]ethan-1-one, BETA-MERCAPTOETHANOL, CHLORIDE ION, ... | | Authors: | Wilson, M.A, Madzelan, P, Rajca, A, Stein, R, Yang, Z. | | Deposit date: | 2022-11-04 | | Release date: | 2023-02-15 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (1.12 Å) | | Cite: | Cucurbit[7]uril Enhances Distance Measurements of Spin-Labeled Proteins.
J.Am.Chem.Soc., 145, 2023
|
|
6OKB
 
 | | Prohead 2 of the phage T5 | | Descriptor: | Major capsid protein | | Authors: | Huet, A, Duda, R.L, Boulanger, P, Conway, J.F. | | Deposit date: | 2019-04-12 | | Release date: | 2019-10-02 | | Last modified: | 2024-03-20 | | Method: | ELECTRON MICROSCOPY (6.7 Å) | | Cite: | Capsid expansion of bacteriophage T5 revealed by high resolution cryoelectron microscopy.
Proc.Natl.Acad.Sci.USA, 116, 2019
|
|
6Q74
 
 | | PI3K delta in complex with 1benzylN[5(3,6dihydro2Hpyran4yl)2methoxypyridin3yl]2methyl1Himidazole4sulfonamide | | Descriptor: | Phosphatidylinositol 4,5-bisphosphate 3-kinase catalytic subunit delta isoform, ~{N}-[5-(3,6-dihydro-2~{H}-pyran-4-yl)-2-methoxy-pyridin-3-yl]-2-methyl-1-(phenylmethyl)imidazole-4-sulfonamide | | Authors: | Convery, M.A, Rowland, P, Down, K, Barton, N. | | Deposit date: | 2018-12-12 | | Release date: | 2018-12-26 | | Last modified: | 2024-06-19 | | Method: | X-RAY DIFFRACTION (2.48 Å) | | Cite: | Discovery of Potent, Efficient, and Selective Inhibitors of Phosphoinositide 3-Kinase delta through a Deconstruction and Regrowth Approach.
J.Med.Chem., 61, 2018
|
|
6Q6Y
 
 | | PI3K delta in complex with N(2chloro5phenylpyridin3yl)benzenesulfonamide | | Descriptor: | Phosphatidylinositol 4,5-bisphosphate 3-kinase catalytic subunit delta isoform, ~{N}-(2-chloranyl-5-phenyl-pyridin-3-yl)benzenesulfonamide | | Authors: | Convery, M.A, Rowland, P, Down, K, Barton, N. | | Deposit date: | 2018-12-12 | | Release date: | 2018-12-26 | | Last modified: | 2024-06-19 | | Method: | X-RAY DIFFRACTION (2.03 Å) | | Cite: | Discovery of Potent, Efficient, and Selective Inhibitors of Phosphoinositide 3-Kinase delta through a Deconstruction and Regrowth Approach.
J.Med.Chem., 61, 2018
|
|
6OMC
 
 | | capsid of T5 virion | | Descriptor: | Major capsid protein | | Authors: | Huet, A, Duda, R.L, Boulanger, P, Conway, J.F. | | Deposit date: | 2019-04-18 | | Release date: | 2019-10-02 | | Last modified: | 2024-03-20 | | Method: | ELECTRON MICROSCOPY (3.8 Å) | | Cite: | Capsid expansion of bacteriophage T5 revealed by high resolution cryoelectron microscopy.
Proc.Natl.Acad.Sci.USA, 116, 2019
|
|
6OMA
 
 | |
6Q73
 
 | | PI3K delta in complex with N[2chloro5(3,6dihydro2Hpyran4yl)pyridin3yl]methanesulfonamide | | Descriptor: | Phosphatidylinositol 4,5-bisphosphate 3-kinase catalytic subunit delta isoform, ~{N}-[2-chloranyl-5-(3,6-dihydro-2~{H}-pyran-4-yl)pyridin-3-yl]methanesulfonamide | | Authors: | Convery, M.A, Rowland, P, Down, K, Barton, N. | | Deposit date: | 2018-12-12 | | Release date: | 2018-12-26 | | Last modified: | 2024-06-19 | | Method: | X-RAY DIFFRACTION (2.21 Å) | | Cite: | Discovery of Potent, Efficient, and Selective Inhibitors of Phosphoinositide 3-Kinase delta through a Deconstruction and Regrowth Approach.
J.Med.Chem., 61, 2018
|
|
6ZAK
 
 | | Room temperature XFEL Isopenicillin N synthase structure in complex with the Fe(IV)=O mimic VO and ACV. | | Descriptor: | Isopenicillin N synthase, L-D-(A-AMINOADIPOYL)-L-CYSTEINYL-D-VALINE, SULFATE ION, ... | | Authors: | Rabe, P, Kamps, J.J.A.G, Sutherlin, K, Pharm, C, McDonough, M.A, Leissing, T.M, Aller, P, Butryn, A, Linyard, J, Lang, P, Brem, J, Fuller, F.D, Batyuk, A, Hunter, M.S, Pettinati, I, Clifton, I.J, Alonso-Mori, R, Gul, S, Young, I, Kim, I, Bhowmick, A, ORiordan, L, Brewster, A.S, Claridge, T.D.W, Sauter, N.K, Yachandra, V, Yano, J, Kern, J.F, Orville, A.M, Schofield, C.J. | | Deposit date: | 2020-06-05 | | Release date: | 2022-01-12 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.54 Å) | | Cite: | Room temperature XFEL Isopenicillin N synthase structure in complex with the Fe(IV)=O mimic VO and ACV.
To Be Published
|
|
4ZFH
 
 | | Crystal structure of Artificial ankyrin repeat protein_Ank(GAG)1D4 mutant -Y56A | | Descriptor: | Artificial ankyrin repeat protein_Ank(GAG)1D4 mutant -Y56A, MAGNESIUM ION | | Authors: | Chuankhayan, P, Saoin, S, Chupradit, K, Wisitponchai, T, Intachai, K, Kitidee, K, Nangola, S, Sanghiran, L.V, Hong, S.S, Boulanger, P, Tayapiwatana, C, Chen, C.J. | | Deposit date: | 2015-04-21 | | Release date: | 2016-04-20 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (1.89 Å) | | Cite: | Crystal structure of Artificial ankyrin repeat protein_Ank(GAG)1D4 mutant- Y56A
To Be Published
|
|
2QWS
 
 | |
