4XMA
 
 | |
4XMI
 
 | |
2IVY
 
 | | Crystal structure of hypothetical protein sso1404 from Sulfolobus solfataricus P2 | | Descriptor: | HYPOTHETICAL PROTEIN SSO1404 | | Authors: | Yan, X, Carter, L.G, Dorward, M, Liu, H, McMahon, S.A, Oke, M, Powers, H, White, M.F, Naismith, J.H. | | Deposit date: | 2006-06-22 | | Release date: | 2006-06-28 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (1.4 Å) | | Cite: | The Scottish Structural Proteomics Facility: Targets, Methods and Outputs.
J.Struct.Funct.Genomics, 11, 2010
|
|
2JG5
 
 | | CRYSTAL STRUCTURE OF A PUTATIVE PHOSPHOFRUCTOKINASE FROM STAPHYLOCOCCUS AUREUS | | Descriptor: | FRUCTOSE 1-PHOSPHATE KINASE | | Authors: | Yan, X, Carter, L.G, Johnson, K.A, Liu, H, Dorward, M, McMahon, S.A, Oke, M, Powers, H, Coote, P.J, Naismith, J.H. | | Deposit date: | 2007-02-08 | | Release date: | 2007-02-27 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | The Scottish Structural Proteomics Facility: Targets, Methods and Outputs.
J.Struct.Funct.Genomics, 11, 2010
|
|
2JG6
 
 | | CRYSTAL STRUCTURE OF A 3-METHYLADENINE DNA GLYCOSYLASE I FROM STAPHYLOCOCCUS AUREUS | | Descriptor: | DNA-3-METHYLADENINE GLYCOSIDASE, ZINC ION | | Authors: | Yan, X, Carter, L.G, Liu, H, Dorward, M, McMahon, S.A, Johnson, K.A, Oke, M, Coote, P.J, Naismith, J.H. | | Deposit date: | 2007-02-08 | | Release date: | 2007-02-20 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | The Scottish Structural Proteomics Facility: Targets, Methods and Outputs.
J.Struct.Funct.Genomics, 11, 2010
|
|
4XJ8
 
 | |
4XE9
 
 | |
4XHB
 
 | |
4XJW
 
 | |
4XOG
 
 | |
1EUU
 
 | | SIALIDASE OR NEURAMINIDASE, LARGE 68KD FORM | | Descriptor: | SIALIDASE, SODIUM ION, beta-D-galactopyranose | | Authors: | Gaskell, A, Crennell, S.J, Taylor, G.L. | | Deposit date: | 1996-06-21 | | Release date: | 1997-01-11 | | Last modified: | 2020-07-29 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | The three domains of a bacterial sialidase: a beta-propeller, an immunoglobulin module and a galactose-binding jelly-roll.
Structure, 3, 1995
|
|
1EUS
 
 | | SIALIDASE COMPLEXED WITH 2-DEOXY-2,3-DEHYDRO-N-ACETYLNEURAMINIC ACID | | Descriptor: | 2-DEOXY-2,3-DEHYDRO-N-ACETYL-NEURAMINIC ACID, SIALIDASE | | Authors: | Gaskell, A, Crennell, S.J, Taylor, G.L. | | Deposit date: | 1996-06-21 | | Release date: | 1997-01-11 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | The three domains of a bacterial sialidase: a beta-propeller, an immunoglobulin module and a galactose-binding jelly-roll.
Structure, 3, 1995
|
|
1EUR
 
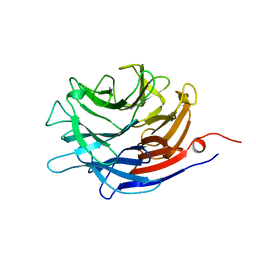 | | SIALIDASE | | Descriptor: | SIALIDASE | | Authors: | Gaskell, A, Crennell, S.J, Taylor, G.L. | | Deposit date: | 1996-06-21 | | Release date: | 1997-01-11 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (1.82 Å) | | Cite: | The three domains of a bacterial sialidase: a beta-propeller, an immunoglobulin module and a galactose-binding jelly-roll.
Structure, 3, 1995
|
|
3SIL
 
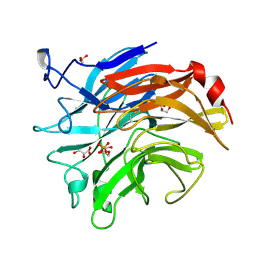 | | SIALIDASE FROM SALMONELLA TYPHIMURIUM | | Descriptor: | GLYCEROL, PHOSPHATE ION, POTASSIUM ION, ... | | Authors: | Garman, E.F, Sheldrick, G.M. | | Deposit date: | 1998-07-07 | | Release date: | 1998-11-11 | | Last modified: | 2023-08-09 | | Method: | X-RAY DIFFRACTION (1.05 Å) | | Cite: | An Enzyme at Atomic Resolution: The 1.05 A Structure of Salmonella Typhimurium Neuraminidase (Sialidase)
To be Published
|
|
1EUT
 
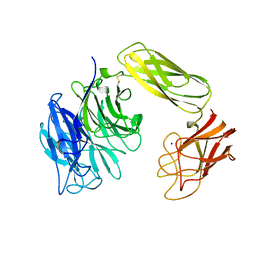 | | SIALIDASE, LARGE 68KD FORM, COMPLEXED WITH GALACTOSE | | Descriptor: | SIALIDASE, SODIUM ION | | Authors: | Gaskell, A, Crennell, S.J, Taylor, G.L. | | Deposit date: | 1996-06-21 | | Release date: | 1997-01-11 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | The three domains of a bacterial sialidase: a beta-propeller, an immunoglobulin module and a galactose-binding jelly-roll.
Structure, 3, 1995
|
|
1E8T
 
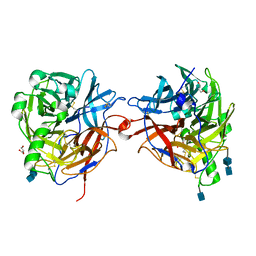 | | Structure of the multifunctional paramyxovirus hemagglutinin-neuraminidase | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, CALCIUM ION, ... | | Authors: | Crennell, S, Takimoto, T, Portner, A, Taylor, G. | | Deposit date: | 2000-10-01 | | Release date: | 2001-04-03 | | Last modified: | 2020-07-29 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Crystal Structure of the Multifunctional Paramyxovirus Hemagglutinin-Neuraminidase
Nat.Struct.Biol., 7, 2000
|
|
1E8V
 
 | | Structure of the multifunctional paramyxovirus hemagglutinin-neuraminidase | | Descriptor: | 2-DEOXY-2,3-DEHYDRO-N-ACETYL-NEURAMINIC ACID, 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Crennell, S, Takimoto, T, Portner, A, Taylor, G. | | Deposit date: | 2000-10-01 | | Release date: | 2001-04-03 | | Last modified: | 2020-07-29 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Crystal Structure of the Multifunctional Paramyxovirus Hemagglutinin-Neuraminidase
Nat.Struct.Biol., 7, 2000
|
|
1E8U
 
 | | Structure of the multifunctional paramyxovirus hemagglutinin-neuraminidase | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, CALCIUM ION, HEMAGGLUTININ-NEURAMINIDASE, ... | | Authors: | Crennell, S, Takimoto, T, Portner, A, Taylor, G. | | Deposit date: | 2000-10-01 | | Release date: | 2001-04-03 | | Last modified: | 2020-07-29 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Crystal Structure of the Multifunctional Paramyxovirus Hemagglutinin-Neuraminidase
Nat.Struct.Biol., 7, 2000
|
|
2X5F
 
 | | Crystal structure of the methicillin-resistant Staphylococcus aureus Sar2028, an aspartate_tyrosine_phenylalanine pyridoxal-5'-phosphate dependent aminotransferase | | Descriptor: | 4-(2-HYDROXYETHYL)-1-PIPERAZINE ETHANESULFONIC ACID, ASPARTATE_TYROSINE_PHENYLALANINE PYRIDOXAL-5' PHOSPHATE-DEPENDENT AMINOTRANSFERASE, MAGNESIUM ION, ... | | Authors: | Oke, M, Carter, L.G, Johnson, K.A, Liu, H, Mcmahon, S.A, White, M.F, Naismith, J.H. | | Deposit date: | 2010-02-08 | | Release date: | 2010-07-21 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | The Scottish Structural Proteomics Facility: Targets, Methods and Outputs.
J.Struct.Funct.Genom., 11, 2010
|
|
2X3F
 
 | | Crystal Structure of the Methicillin-Resistant Staphylococcus aureus Sar2676, a Pantothenate Synthetase. | | Descriptor: | DIPHOSPHOMETHYLPHOSPHONIC ACID ADENOSYL ESTER, PANTHOTHENATE SYNTHETASE, SULFATE ION | | Authors: | Oke, M, Carter, L.G, Johnson, K.A, Liu, H, Mcmahon, S.A, White, M.F, Naismith, J.H. | | Deposit date: | 2010-01-24 | | Release date: | 2010-07-21 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | The Scottish Structural Proteomics Facility: Targets, Methods and Outputs.
J.Struct.Funct.Genom., 11, 2010
|
|
2X0O
 
 | | Apo structure of the Alcaligin biosynthesis protein C (AlcC) from Bordetella bronchiseptica | | Descriptor: | ALCALIGIN BIOSYNTHESIS PROTEIN, SULFATE ION | | Authors: | Johnson, K.A, Schmelz, S, Kadi, N, Mcmahon, S.A, Oke, M, Liu, H, Carter, L.G, White, M.F, Challis, G.L, Naismith, J.H. | | Deposit date: | 2009-12-16 | | Release date: | 2010-07-28 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | The Scottish Structural Proteomics Facility: Targets, Methods and Outputs.
J.Struct.Funct.Genomics, 11, 2010
|
|
4FP2
 
 | |
4FPL
 
 | |
4FOV
 
 | |
4FPC
 
 | |
