3EDF
 
 | | Structural base for cyclodextrin hydrolysis | | Descriptor: | CALCIUM ION, Cyclohexakis-(1-4)-(alpha-D-glucopyranose), Cyclomaltodextrinase, ... | | Authors: | Buedenbender, S, Schulz, G.E. | | Deposit date: | 2008-09-03 | | Release date: | 2009-03-03 | | Last modified: | 2024-05-29 | | Method: | X-RAY DIFFRACTION (1.65 Å) | | Cite: | Structural base for enzymatic cyclodextrin hydrolysis
J.Mol.Biol., 385, 2009
|
|
3EDJ
 
 | | Structural base for cyclodextrin hydrolysis | | Descriptor: | CALCIUM ION, Cycloheptakis-(1-4)-(alpha-D-glucopyranose), Cyclomaltodextrinase, ... | | Authors: | Buedenbender, S, Schulz, G.E. | | Deposit date: | 2008-09-03 | | Release date: | 2009-03-03 | | Last modified: | 2024-05-29 | | Method: | X-RAY DIFFRACTION (1.69 Å) | | Cite: | Structural base for enzymatic cyclodextrin hydrolysis
J.Mol.Biol., 385, 2009
|
|
3EDE
 
 | |
3EDK
 
 | | Structural base for cyclodextrin hydrolysis | | Descriptor: | CALCIUM ION, Cyclomaltodextrinase, Cyclooctakis-(1-4)-(alpha-D-glucopyranose), ... | | Authors: | Buedenbender, S, Schulz, G.E. | | Deposit date: | 2008-09-03 | | Release date: | 2009-03-03 | | Last modified: | 2024-05-29 | | Method: | X-RAY DIFFRACTION (1.77 Å) | | Cite: | Structural base for enzymatic cyclodextrin hydrolysis
J.Mol.Biol., 385, 2009
|
|
3EDD
 
 | | Structural base for cyclodextrin hydrolysis | | Descriptor: | CALCIUM ION, Cyclohexakis-(1-4)-(alpha-D-glucopyranose), Cyclomaltodextrinase | | Authors: | Buedenbender, S, Schulz, G.E. | | Deposit date: | 2008-09-03 | | Release date: | 2009-03-03 | | Last modified: | 2024-05-29 | | Method: | X-RAY DIFFRACTION (2.65 Å) | | Cite: | Structural base for enzymatic cyclodextrin hydrolysis
J.Mol.Biol., 385, 2009
|
|
1NKS
 
 | |
1MPR
 
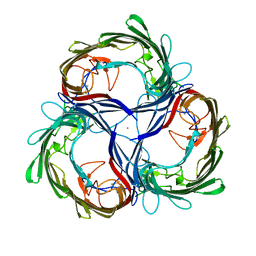 | | MALTOPORIN FROM SALMONELLA TYPHIMURIUM | | Descriptor: | CALCIUM ION, MALTOPORIN | | Authors: | Meyer, J.E.W, Schulz, G.E. | | Deposit date: | 1996-12-18 | | Release date: | 1997-03-12 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Structure of maltoporin from Salmonella typhimurium ligated with a nitrophenyl-maltotrioside.
J.Mol.Biol., 266, 1997
|
|
2CB6
 
 | | Crystal structure of the catalytic domain of the mosquitocidal toxin from Bacillus sphaericus, mutant E195Q | | Descriptor: | MOSQUITOCIDAL TOXIN | | Authors: | Reinert, D.J, Carpusca, I, Aktories, K, Schulz, G.E. | | Deposit date: | 2005-12-29 | | Release date: | 2006-02-22 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | Structure of the Mosquitocidal Toxin from Bacillus Sphaericus.
J.Mol.Biol., 357, 2006
|
|
2CB4
 
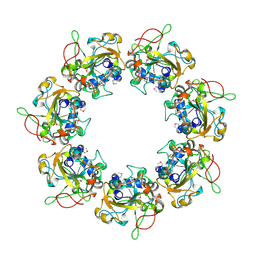 | | Crystal structure of the catalytic domain of the mosquitocidal toxin from Bacillus sphaericus, mutant E197Q | | Descriptor: | MOSQUITOCIDAL TOXIN | | Authors: | Reinert, D.J, Carpusca, I, Aktories, K, Schulz, G.E. | | Deposit date: | 2005-12-29 | | Release date: | 2006-02-22 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Structure of the Mosquitocidal Toxin from Bacillus Sphaericus.
J.Mol.Biol., 357, 2006
|
|
2F5V
 
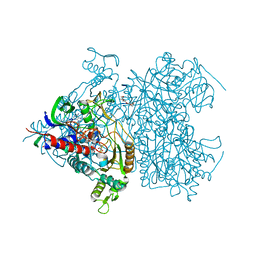 | | Reaction geometry and thermostability mutant of pyranose 2-oxidase from the white-rot fungus Peniophora sp. | | Descriptor: | DI(HYDROXYETHYL)ETHER, FLAVIN-ADENINE DINUCLEOTIDE, Pyranose 2-oxidase, ... | | Authors: | Bannwarth, M, Bastian, S, Heckmann-Pohl, D, Giffhorn, F, Schulz, G.E. | | Deposit date: | 2005-11-28 | | Release date: | 2006-06-13 | | Last modified: | 2024-10-09 | | Method: | X-RAY DIFFRACTION (1.41 Å) | | Cite: | Reaction Geometry and Thermostable Variant of Pyranose 2-Oxidase from the White-Rot Fungus Peniophora sp.
Biochemistry, 45, 2006
|
|
2F6C
 
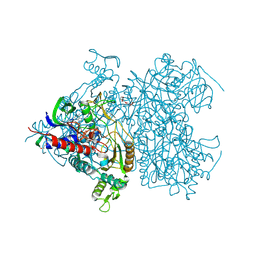 | | Reaction geometry and thermostability of pyranose 2-oxidase from the white-rot fungus Peniophora sp., Thermostability mutant E542K | | Descriptor: | DI(HYDROXYETHYL)ETHER, FLAVIN-ADENINE DINUCLEOTIDE, Pyranose 2-oxidase, ... | | Authors: | Bannwarth, M, Heckmann-Pohl, D.M, Bastian, S, Giffhorn, F, Schulz, G.E. | | Deposit date: | 2005-11-29 | | Release date: | 2006-06-13 | | Last modified: | 2024-11-20 | | Method: | X-RAY DIFFRACTION (1.84 Å) | | Cite: | Reaction Geometry and Thermostable Variant of Pyranose 2-Oxidase from the White-Rot Fungus Peniophora sp.
Biochemistry, 45, 2006
|
|
1O9H
 
 | |
1O9G
 
 | |
1OG4
 
 | | Crystal Structure of the Eucaryotic Mono-ADP-Ribosyltransferase ART2.2 Mutant E189A in Complex with NADH | | Descriptor: | 1,4-DIHYDRONICOTINAMIDE ADENINE DINUCLEOTIDE, T-CELL ECTO-ADP-RIBOSYLTRANSFERASE 2 | | Authors: | Ritter, H, Koch-Nolte, F, Marquez, V.E, Schulz, G.E. | | Deposit date: | 2003-04-24 | | Release date: | 2003-08-28 | | Last modified: | 2024-11-13 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Substrate Binding and Catalysis of Ecto-Adp-Ribosyltransferase 2.2 From Rat
Biochemistry, 42, 2003
|
|
1OKX
 
 | | Binding Structure of Elastase Inhibitor Scyptolin A | | Descriptor: | ELASTASE 1, SCYPTOLIN A | | Authors: | Matern, U, Schleberger, C, Jelakovic, S, Weckesser, J, Schulz, G.E. | | Deposit date: | 2003-07-31 | | Release date: | 2003-10-24 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Binding Structure of Elastase Inhibitor Scyptolin A
Chem.Biol., 10, 2003
|
|
1OG3
 
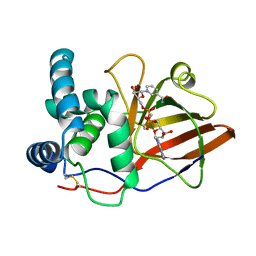 | | Crystal structure of the eukaryotic mono-ADP-ribosyltransferase ART2.2 mutant E189I in complex with NAD | | Descriptor: | NICOTINAMIDE-ADENINE-DINUCLEOTIDE, T-CELL ECTO-ADP-RIBOSYLTRANSFERASE 2 | | Authors: | Ritter, H, Koch-Nolte, F, Marquez, V.E, Schulz, G.E. | | Deposit date: | 2003-04-24 | | Release date: | 2003-08-28 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Substrate Binding and Catalysis of Ecto-Adp-Ribosyltransferase 2.2 From Rat
Biochemistry, 42, 2003
|
|
1OG1
 
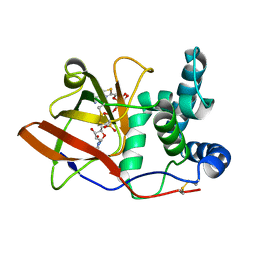 | | CRYSTAL STRUCTURE OF THE EUCARYOTIC MONO-ADP-RIBOSYLTRANSFERASE ART2.2 IN COMPLEX WITH TAD | | Descriptor: | BETA-METHYLENE-THIAZOLE-4-CARBOXYAMIDE-ADENINE DINUCLEOTIDE, T-CELL ECTO-ADP-RIBOSYLTRANSFERASE 2 | | Authors: | Ritter, H, Koch-Nolte, F, Marquez, V.E, Schulz, G.E. | | Deposit date: | 2003-04-23 | | Release date: | 2003-08-28 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Substrate Binding and Catalysis of Ecto-Adp-Ribosyltransferase 2.2 From Rat
Biochemistry, 42, 2003
|
|
1UWK
 
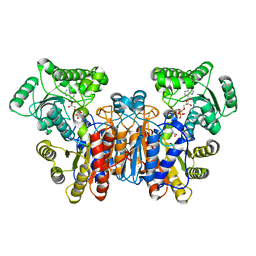 | |
1X7P
 
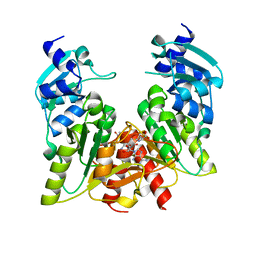 | |
1W27
 
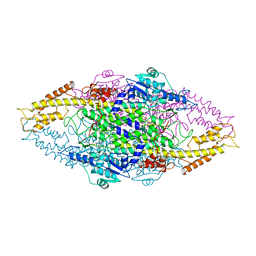 | | Phenylalanine ammonia-lyase (PAL) from Petroselinum crispum | | Descriptor: | 2,3-DIHYDROXY-1,4-DITHIOBUTANE, PHENYLALANINE AMMONIA-LYASE 1 | | Authors: | Ritter, H, Schulz, G.E. | | Deposit date: | 2004-06-29 | | Release date: | 2004-11-25 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Structural Basis for the Entrance Into the Phenylpropanoid Metabolism Catalyzed by Phenylalanine Ammonia-Lyase
Plant Cell, 16, 2004
|
|
1W1U
 
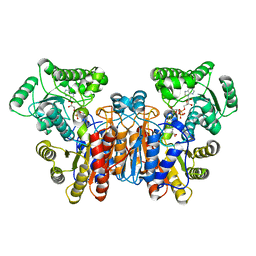 | |
1UUN
 
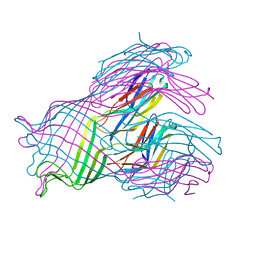 | |
1UKZ
 
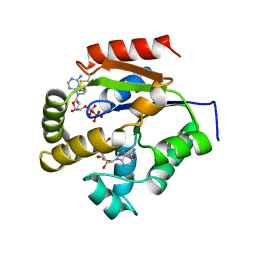 | |
1X7O
 
 | |
1W4R
 
 | | Structure of a type II thymidine kinase with bound dTTP | | Descriptor: | (2R,3S)-1,4-DIMERCAPTOBUTANE-2,3-DIOL, THYMIDINE KINASE, THYMIDINE-5'-TRIPHOSPHATE, ... | | Authors: | Birringer, M.S, Claus, M.T, Folkers, G, Kloer, D.P, Schulz, G.E, Scapozza, L. | | Deposit date: | 2004-07-27 | | Release date: | 2005-02-01 | | Last modified: | 2025-04-09 | | Method: | X-RAY DIFFRACTION (1.83 Å) | | Cite: | Structure of a Type II Thymidine Kinase with Bound Dttp
FEBS Lett., 579, 2005
|
|
