1JIB
 
 | | Complex of Alpha-amylase II (TVA II) from Thermoactinomyces vulgaris R-47 with Maltotetraose Based on a Crystal Soaked with Maltohexaose. | | Descriptor: | NEOPULLULANASE, alpha-D-glucopyranose-(1-4)-alpha-D-glucopyranose-(1-4)-alpha-D-glucopyranose-(1-4)-alpha-D-glucopyranose | | Authors: | Yokota, T, Tonozuka, T, Shimura, Y, Ichikawa, K, Kamitori, S, Sakano, Y. | | Deposit date: | 2001-07-02 | | Release date: | 2001-07-25 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (3.3 Å) | | Cite: | Structures of Thermoactinomyces vulgaris R-47 alpha-amylase II complexed with substrate analogues.
Biosci.Biotechnol.Biochem., 65, 2001
|
|
1WKI
 
 | | solution structure of ribosomal protein L16 from thermus thermophilus HB8 | | Descriptor: | LSU ribosomal protein L16P | | Authors: | Nishimura, M, Yoshida, T, Shirouzu, M, Terada, T, Kuramitsu, S, Yokoyama, S, Ohkubo, T, Kobayashi, Y, RIKEN Structural Genomics/Proteomics Initiative (RSGI) | | Deposit date: | 2004-05-31 | | Release date: | 2004-12-14 | | Last modified: | 2024-05-01 | | Method: | SOLUTION NMR | | Cite: | Solution Structure of Ribosomal Protein L16 from Thermus thermophilus HB8
J.Mol.Biol., 344, 2004
|
|
6AKG
 
 | |
6AKE
 
 | |
3WVS
 
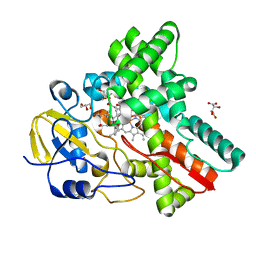 | | Crystal Structure of Cytochrome P450revI | | Descriptor: | (2E,4S,5S,6E,8E)-10-{(2R,3S,6S,8R,9S)-9-butyl-8-[(1E,3E)-4-carboxy-3-methylbuta-1,3-dien-1-yl]-3-methyl-1,7-dioxaspiro[5.5]undec-2-yl}-5-hydroxy-4,8-dimethyldeca-2,6,8-trienoic acid, GLYCEROL, L(+)-TARTARIC ACID, ... | | Authors: | Nagano, S, Takahashi, S, Osada, H, Shiro, Y. | | Deposit date: | 2014-06-06 | | Release date: | 2014-10-01 | | Last modified: | 2024-05-29 | | Method: | X-RAY DIFFRACTION (1.4 Å) | | Cite: | Structure-function analyses of cytochrome P450revI involved in reveromycin A biosynthesis and evaluation of the biological activity of its substrate, reveromycin T.
J.Biol.Chem., 289, 2014
|
|
1FQY
 
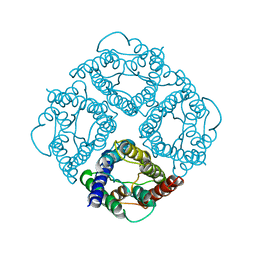 | | STRUCTURE OF AQUAPORIN-1 AT 3.8 A RESOLUTION BY ELECTRON CRYSTALLOGRAPHY | | Descriptor: | AQUAPORIN-1 | | Authors: | Murata, K, Mitsuoka, K, Hirai, T, Walz, T, Agre, P, Heymann, J.B, Engel, A, Fujiyoshi, Y. | | Deposit date: | 2000-09-07 | | Release date: | 2000-10-18 | | Last modified: | 2024-04-17 | | Method: | ELECTRON CRYSTALLOGRAPHY (3.8 Å) | | Cite: | Structural determinants of water permeation through aquaporin-1.
Nature, 407, 2000
|
|
1GE6
 
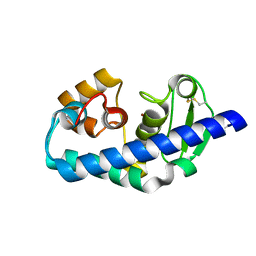 | | ZINC PEPTIDASE FROM GRIFOLA FRONDOSA | | Descriptor: | PEPTIDYL-LYS METALLOENDOPEPTIDASE, ZINC ION, alpha-D-mannopyranose | | Authors: | Hori, T, Kumasaka, T, Yamamoto, M, Nonaka, T, Tanaka, N, Hashimoto, Y, Ueki, T, Takio, K. | | Deposit date: | 2000-10-11 | | Release date: | 2001-03-14 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Structure of a new 'aspzincin' metalloendopeptidase from Grifola frondosa: implications for the catalytic mechanism and substrate specificity based on several different crystal forms.
Acta Crystallogr.,Sect.D, 57, 2001
|
|
1OED
 
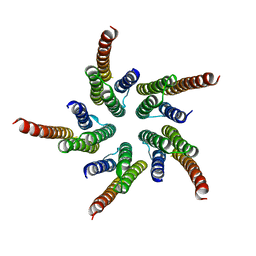 | | STRUCTURE OF ACETYLCHOLINE RECEPTOR PORE FROM ELECTRON IMAGES | | Descriptor: | Acetylcholine receptor beta subunit, Acetylcholine receptor delta subunit, Acetylcholine receptor gamma subunit, ... | | Authors: | Miyazawa, A, Fujiyoshi, Y, Unwin, N. | | Deposit date: | 2003-03-24 | | Release date: | 2003-06-26 | | Last modified: | 2024-05-08 | | Method: | ELECTRON MICROSCOPY (4 Å) | | Cite: | Structure and Gating Mechanism of the Acetylcholine Receptor Pore.
Nature, 423, 2003
|
|
1GE5
 
 | | ZINC PEPTIDASE FROM GRIFOLA FRONDOSA | | Descriptor: | PEPTIDYL-LYS METALLOENDOPEPTIDASE, ZINC ION, alpha-D-mannopyranose | | Authors: | Hori, T, Kumasaka, T, Yamamoto, M, Nonaka, T, Tanaka, N, Hashimoto, Y, Ueki, T, Takio, K. | | Deposit date: | 2000-10-11 | | Release date: | 2001-03-14 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Structure of a new 'aspzincin' metalloendopeptidase from Grifola frondosa: implications for the catalytic mechanism and substrate specificity based on several different crystal forms.
Acta Crystallogr.,Sect.D, 57, 2001
|
|
1G12
 
 | | ZINC PEPTIDASE FROM GRIFOLA FRONDOSA | | Descriptor: | PEPTIDYL-LYS METALLOENDOPEPTIDASE, ZINC ION, alpha-D-mannopyranose | | Authors: | Hori, T, Kumasaka, T, Yamamoto, M, Nonaka, T, Tanaka, N, Hashimoto, Y, Ueki, T, Takio, K. | | Deposit date: | 2000-10-10 | | Release date: | 2001-03-14 | | Last modified: | 2020-07-29 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Structure of a new 'aspzincin' metalloendopeptidase from Grifola frondosa: implications for the catalytic mechanism and substrate specificity based on several different crystal forms.
Acta Crystallogr.,Sect.D, 57, 2001
|
|
1GED
 
 | | A positive charge route for the access of nadh to heme formed in the distal heme pocket of cytochrome p450nor | | Descriptor: | BROMIDE ION, CYTOCHROME P450 55A1, PROTOPORPHYRIN IX CONTAINING FE | | Authors: | Kudo, T, Takaya, N, Park, S.-Y, Shiro, Y, Shoun, H. | | Deposit date: | 2000-11-02 | | Release date: | 2000-11-22 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | A positively charged cluster formed in the heme-distal pocket of cytochrome P450nor is essential for interaction with NADH
J.Biol.Chem., 276, 2001
|
|
1GE7
 
 | | ZINC PEPTIDASE FROM GRIFOLA FRONDOSA | | Descriptor: | PEPTIDYL-LYS METALLOENDOPEPTIDASE, ZINC ION, alpha-D-mannopyranose | | Authors: | Hori, T, Kumasaka, T, Yamamoto, M, Nonaka, T, Tanaka, N, Hashimoto, Y, Ueki, T, Takio, K. | | Deposit date: | 2000-10-11 | | Release date: | 2001-03-14 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Structure of a new 'aspzincin' metalloendopeptidase from Grifola frondosa: implications for the catalytic mechanism and substrate specificity based on several different crystal forms.
Acta Crystallogr.,Sect.D, 57, 2001
|
|
1GE8
 
 | |
1IY3
 
 | | Solution Structure of the Human lysozyme at 4 degree C | | Descriptor: | Lysozyme | | Authors: | Kumeta, H, Miura, A, Kobashigawa, Y, Miura, K, Oka, C, Nitta, K, Nemoto, N, Tsuda, S. | | Deposit date: | 2002-07-15 | | Release date: | 2002-07-31 | | Last modified: | 2023-12-27 | | Method: | SOLUTION NMR | | Cite: | Low-temperature-induced structural changes in human lysozyme elucidated by three-dimensional NMR spectroscopy
Biochemistry, 42, 2003
|
|
1IO9
 
 | | THERMOPHILIC CYTOCHROME P450 (CYP119) FROM SULFOLOBUS SOLFATARICUS: HIGH RESOLUTION STRUCTURAL ORIGIN OF ITS THERMOSTABILITY AND FUNCTIONAL PROPERTIES | | Descriptor: | CYTOCHROME P450 CYP119, PROTOPORPHYRIN IX CONTAINING FE | | Authors: | Park, S.-Y, Yamane, K, Adachi, S, Shiro, Y, Sligar, S.G, RIKEN Structural Genomics/Proteomics Initiative (RSGI) | | Deposit date: | 2001-02-08 | | Release date: | 2001-03-07 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (2.05 Å) | | Cite: | Thermophilic cytochrome P450 (CYP119) from Sulfolobus solfataricus: high resolution structure and functional properties.
J.Inorg.Biochem., 91, 2002
|
|
1J2G
 
 | |
1IO8
 
 | | Thermophilic cytochrome P450 (CYP119) from sulfolobus solfataricus: High resolution structural origin of its thermostability and functional properties | | Descriptor: | CYTOCHROME P450 CYP119, PROTOPORPHYRIN IX CONTAINING FE | | Authors: | Park, S.-Y, Yamane, K, Adachi, S, Shiro, Y, Sligar, S.G, RIKEN Structural Genomics/Proteomics Initiative (RSGI) | | Deposit date: | 2001-02-08 | | Release date: | 2001-03-07 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Thermophilic cytochrome P450 (CYP119) from Sulfolobus solfataricus: high resolution structure and functional properties
J.Inorg.Biochem., 91, 2002
|
|
1IR8
 
 | | IM mutant of lysozyme | | Descriptor: | lysozyme | | Authors: | Ohmura, T, Ueda, T, Hashimoto, Y, Imoto, T. | | Deposit date: | 2001-09-19 | | Release date: | 2001-10-03 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (1.63 Å) | | Cite: | Tolerance of point substitution of methionine for isoleucine in hen egg white lysozyme.
Protein Eng., 14, 2001
|
|
1IY4
 
 | | Solution structure of the human lysozyme at 35 degree C | | Descriptor: | Lysozyme | | Authors: | Kumeta, H, Miura, A, Kobashigawa, Y, Miura, K, Oka, C, Nitta, K, Nemoto, N, Tsuda, S. | | Deposit date: | 2002-07-15 | | Release date: | 2002-07-31 | | Last modified: | 2023-12-27 | | Method: | SOLUTION NMR | | Cite: | Low-temperature-induced structural changes in human lysozyme elucidated by three-dimensional NMR spectroscopy
Biochemistry, 42, 2003
|
|
8YFY
 
 | | CRYSTAL STRUCTURE OF THE EST1 H274D MUTANT AT PH 4.2 | | Descriptor: | Carboxylesterase, octyl beta-D-glucopyranoside | | Authors: | Unno, H, Oshima, Y, Nishino, T, Nakayama, T, Kusunoki, M. | | Deposit date: | 2024-02-26 | | Release date: | 2024-07-10 | | Last modified: | 2024-08-28 | | Method: | X-RAY DIFFRACTION (1.45 Å) | | Cite: | Lowering pH optimum of activity of SshEstI, a slightly alkaliphilic archaeal esterase of the hormone-sensitive lipase family.
J.Biosci.Bioeng., 138, 2024
|
|
8YFZ
 
 | | CRYSTAL STRUCTURE OF THE EST1 H274E MUTANT AT PH 4.2 | | Descriptor: | Carboxylesterase, octyl beta-D-glucopyranoside | | Authors: | Unno, H, Oshima, Y, Nishino, T, Nakayama, T, Kusunoki, M. | | Deposit date: | 2024-02-26 | | Release date: | 2024-07-10 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Lowering pH optimum of activity of SshEstI, a slightly alkaliphilic archaeal esterase of the hormone-sensitive lipase family.
J.Biosci.Bioeng., 2024
|
|
1TMW
 
 | | Solution structure of Human Coactosin Like Protein D123N | | Descriptor: | Coactosin-like protein | | Authors: | Dai, H, Wu, J, Xu, Y, Tang, Y, Ding, H, Shi, Y. | | Deposit date: | 2004-06-11 | | Release date: | 2005-06-28 | | Last modified: | 2024-05-29 | | Method: | SOLUTION NMR | | Cite: | Study on Solution Structure and Its binding function to F-actin
To be Published
|
|
5B66
 
 | | Crystal structure analysis of Photosystem II complex | | Descriptor: | (3R)-beta,beta-caroten-3-ol, 1,2-DI-O-ACYL-3-O-[6-DEOXY-6-SULFO-ALPHA-D-GLUCOPYRANOSYL]-SN-GLYCEROL, 1,2-DIPALMITOYL-PHOSPHATIDYL-GLYCEROLE, ... | | Authors: | Tanaka, A, Fukushima, Y, Kamiya, N. | | Deposit date: | 2016-05-25 | | Release date: | 2017-02-01 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | Two Different Structures of the Oxygen-Evolving Complex in the Same Polypeptide Frameworks of Photosystem II
J. Am. Chem. Soc., 139, 2017
|
|
1U4A
 
 | | Solution structure of human SUMO-3 C47S | | Descriptor: | Ubiquitin-like protein SMT3A | | Authors: | Ding, H, Xu, Y, Dai, H, Tang, Y, Wu, J, Shi, Y. | | Deposit date: | 2004-07-23 | | Release date: | 2005-03-08 | | Last modified: | 2024-05-29 | | Method: | SOLUTION NMR | | Cite: | Solution Structure of Human SUMO-3 C47S and Its Binding Surface for Ubc9
Biochemistry, 44, 2005
|
|
1UG2
 
 | | Solution Structure of Mouse Hypothetical Gene (2610100B20Rik) Product Homologous to Myb DNA-binding Domain | | Descriptor: | 2610100B20Rik gene product | | Authors: | Zhao, C, Kigawa, T, Tochio, N, Koshiba, S, Inoue, M, Shirouzu, M, Terada, T, Yabuki, T, Aoki, M, Seki, E, Matsuda, T, Tanaka, A, Osanai, T, Arakawa, T, Carninci, P, Kawai, J, Hayashizaki, Y, Yokoyama, S, RIKEN Structural Genomics/Proteomics Initiative (RSGI) | | Deposit date: | 2003-06-11 | | Release date: | 2004-06-22 | | Last modified: | 2023-12-27 | | Method: | SOLUTION NMR | | Cite: | Solution Structure of Mouse Hypothetical Gene (2610100B20Rik) Product Homologous to Myb DNA-binding Domain
To be Published
|
|
