6AWK
 
 | |
6AZG
 
 | |
3FGQ
 
 | | Crystal structure of native human neuroserpin | | Descriptor: | GLYCEROL, Neuroserpin | | Authors: | Takehara, S, Yang, X, Mikami, B, Onda, M. | | Deposit date: | 2008-12-08 | | Release date: | 2009-04-28 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (2.09 Å) | | Cite: | The 2.1-A crystal structure of native neuroserpin reveals unique structural elements that contribute to conformational instability
J.Mol.Biol., 388, 2009
|
|
3FHO
 
 | | Structure of S. pombe Dbp5 | | Descriptor: | ATP-dependent RNA helicase dbp5 | | Authors: | Cheng, Z, Song, H. | | Deposit date: | 2008-12-09 | | Release date: | 2009-10-13 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Solution and crystal structures of mRNA exporter Dbp5p and its interaction with nucleotides
J.Mol.Biol., 388, 2009
|
|
6AXI
 
 | | PawL-Derived Peptide PLP-2 | | Descriptor: | ASP-LEU-PHE-VAL-PRO-PRO-ILE-ASP | | Authors: | Fisher, M, Mylne, J.S, Howard, M.J. | | Deposit date: | 2017-09-06 | | Release date: | 2018-03-07 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | A family of small, cyclic peptides buried in preproalbumin since the Eocene epoch.
Plant Direct, 2, 2018
|
|
6BHX
 
 | | B. subtilis SsbA with DNA | | Descriptor: | DNA (5'-D(P*TP*TP*TP*TP*TP*TP*TP*TP*TP*TP*T)-3'), Single-stranded DNA-binding protein A | | Authors: | Dubiel, K.D, Myers, A.R, Satyshur, K.A, Keck, J.L. | | Deposit date: | 2017-10-31 | | Release date: | 2018-12-19 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (2.936 Å) | | Cite: | Structural Mechanisms of Cooperative DNA Binding by Bacterial Single-Stranded DNA-Binding Proteins.
J. Mol. Biol., 431, 2019
|
|
6BHW
 
 | | B. subtilis SsbA | | Descriptor: | 1,2-ETHANEDIOL, DI(HYDROXYETHYL)ETHER, Single-stranded DNA-binding protein A | | Authors: | Dubiel, K.D, Myers, A.R, Satyshur, K.A, Keck, J.L. | | Deposit date: | 2017-10-31 | | Release date: | 2018-12-19 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (2.208 Å) | | Cite: | Structural Mechanisms of Cooperative DNA Binding by Bacterial Single-Stranded DNA-Binding Proteins.
J. Mol. Biol., 431, 2019
|
|
6AXJ
 
 | |
3GK7
 
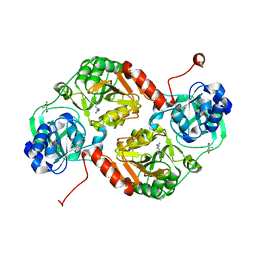 | | Crystal structure of 4-hydroxybutyrate CoA-Transferase from Clostridium aminobutyricum | | Descriptor: | 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, 4-Hydroxybutyrate CoA-transferase, SPERMIDINE | | Authors: | Messerschmidt, A, Macieira, S, Velarde, M. | | Deposit date: | 2009-03-10 | | Release date: | 2009-12-22 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | Crystal structure of 4-hydroxybutyrate CoA-transferase from Clostridium aminobutyricum
Biol.Chem., 390, 2009
|
|
7KW1
 
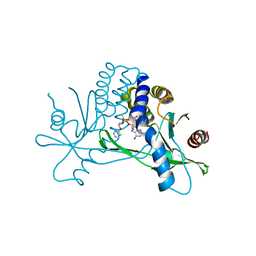 | | Structure of hSTING in complex with novel carbocyclic pyrimidine CDN-3 | | Descriptor: | (2R,5R,7R,8R,10R,12aR,14R,15aS,16R)-7-(2-amino-6-oxo-1,6-dihydro-9H-purin-9-yl)-16-hydroxy-14-[(pyrimidin-4-yl)oxy]-2,10-disulfanyldecahydro-2H,10H-5,8-methano-2lambda~5~,10lambda~5~-cyclopenta[l][1,3,6,9,11,2,10]pentaoxadiphosphacyclotetradecine-2,10-dione, Stimulator of interferon genes protein | | Authors: | Skene, R. | | Deposit date: | 2020-11-29 | | Release date: | 2021-06-02 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Identification of Novel Carbocyclic Pyrimidine Cyclic Dinucleotide STING Agonists for Antitumor Immunotherapy Using Systemic Intravenous Route.
J.Med.Chem., 64, 2021
|
|
7KVX
 
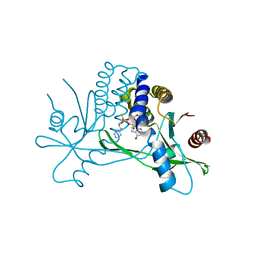 | | Structure of hSTING in complex with novel carbocyclic pyrimidine CDN 1 | | Descriptor: | (2R,5R,7R,8R,10R,12aR,14R,15aS,16R)-7-(2-amino-6-oxo-1,6-dihydro-9H-purin-9-yl)-16-hydroxy-14-[(pyrimidin-4-yl)amino]-2,10-disulfanyldecahydro-2H,10H-5,8-methano-2lambda~5~,10lambda~5~-cyclopenta[l][1,3,6,9,11,2,10]pentaoxadiphosphacyclotetradecine-2,10-dione, Stimulator of interferon genes protein | | Authors: | Skene, R. | | Deposit date: | 2020-11-29 | | Release date: | 2021-06-02 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (2.48 Å) | | Cite: | Identification of Novel Carbocyclic Pyrimidine Cyclic Dinucleotide STING Agonists for Antitumor Immunotherapy Using Systemic Intravenous Route.
J.Med.Chem., 64, 2021
|
|
7KVZ
 
 | | Structure of hSTING in complex with novel carbocyclic pyrimidine CDN-2 | | Descriptor: | (2R,5R,7R,8R,10S,12aR,14R,15aS,16R)-7-(2-amino-6-oxo-1,6-dihydro-9H-purin-9-yl)-2,10,16-trihydroxy-14-[(pyrimidin-4-yl)oxy]decahydro-2H,10H-5,8-methano-2lambda~5~,10lambda~5~-cyclopenta[l][1,3,6,9,11,2,10]pentaoxadiphosphacyclotetradecine-2,10-dione, Stimulator of interferon genes protein | | Authors: | Skene, R.J. | | Deposit date: | 2020-11-29 | | Release date: | 2022-02-09 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (2.35 Å) | | Cite: | Identification of Novel Carbocyclic Pyrimidine Cyclic Dinucleotide STING Agonists for Antitumor Immunotherapy Using Systemic Intravenous Route.
J.Med.Chem., 64, 2021
|
|
8ETN
 
 | |
5WF0
 
 | |
5WFK
 
 | |
5Y1U
 
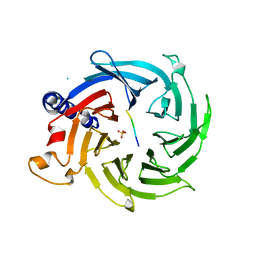 | | Crystal structure of RBBP4 bound to AEBP2 RRK motif | | Descriptor: | Histone-binding protein RBBP4, SULFATE ION, Zinc finger protein AEBP2 | | Authors: | Sun, A, Li, F, Wu, J, Shi, Y. | | Deposit date: | 2017-07-21 | | Release date: | 2018-04-18 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.141 Å) | | Cite: | Structural and biochemical insights into human zinc finger protein AEBP2 reveals interactions with RBBP4
Protein Cell, 2017
|
|
5YJH
 
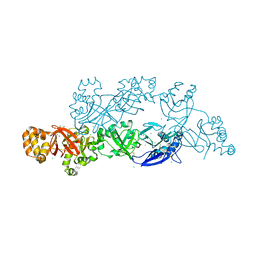 | | Structural insights into periostin functions | | Descriptor: | CALCIUM ION, CHLORIDE ION, MAGNESIUM ION, ... | | Authors: | Liu, H, Liu, J, Xu, F. | | Deposit date: | 2017-10-10 | | Release date: | 2018-05-23 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.957 Å) | | Cite: | Structural characterizations of human periostin dimerization and cysteinylation.
FEBS Lett., 592, 2018
|
|
6MIN
 
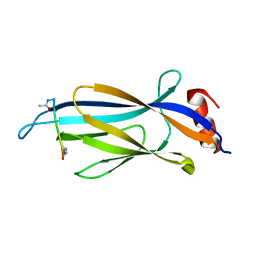 | |
6MIM
 
 | |
6MIO
 
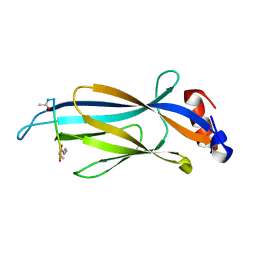 | | Crystal structure of Taf14 YEATS domain in complex with histone H3K9pr | | Descriptor: | Histone H3K9pr, Transcription initiation factor TFIID subunit 14 | | Authors: | Klein, B.J, Andrews, F.H, Vann, K.R, Kutateladze, T.G. | | Deposit date: | 2018-09-19 | | Release date: | 2018-11-14 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | Structural insights into the pi-pi-pi stacking mechanism and DNA-binding activity of the YEATS domain.
Nat Commun, 9, 2018
|
|
7VWA
 
 | |
7VW8
 
 | |
7VW9
 
 | |
6MIP
 
 | |
5YJG
 
 | | Structural insights into periostin functions | | Descriptor: | CALCIUM ION, CHLORIDE ION, CYSTEINE, ... | | Authors: | Liu, H, Liu, J, Xu, F. | | Deposit date: | 2017-10-10 | | Release date: | 2018-05-23 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.399 Å) | | Cite: | Structural characterizations of human periostin dimerization and cysteinylation.
FEBS Lett., 592, 2018
|
|
