6O19
 
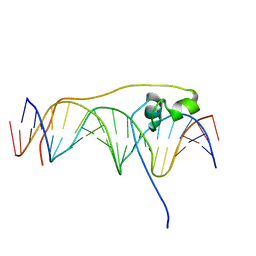 | | Crystal Structure of Pho7 complex with pho1 promoter site 2 | | Descriptor: | DNA (5'-D(*GP*TP*TP*TP*TP*TP*AP*AP*TP*TP*TP*CP*CP*GP*AP*AP*TP*AP*AP*T)-3'), DNA (5'-D(*TP*TP*AP*TP*TP*CP*GP*GP*AP*AP*AP*TP*TP*AP*AP*AP*AP*AP*CP*A)-3'), Transcription factor Pho7, ... | | Authors: | Garg, A, Goldgur, Y, Shuman, S. | | Deposit date: | 2019-02-18 | | Release date: | 2019-04-24 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (1.596 Å) | | Cite: | Structure of Fission Yeast Transcription Factor Pho7 Bound topho1Promoter DNA and Effect of Pho7 Mutations on DNA Binding and Phosphate Homeostasis.
Mol.Cell.Biol., 39, 2019
|
|
8DCD
 
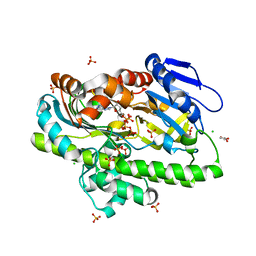 | | RNA ligase RtcB from Pyrococcus horikoshii in complex with Zn2+ and GTP | | Descriptor: | CHLORIDE ION, GLYCEROL, GUANOSINE-5'-TRIPHOSPHATE, ... | | Authors: | Jacewicz, A, Dantuluri, S, Shuman, S. | | Deposit date: | 2022-06-16 | | Release date: | 2022-10-12 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (2.22 Å) | | Cite: | Structures of RNA ligase RtcB in complexes with divalent cations and GTP.
Rna, 28, 2022
|
|
8DC9
 
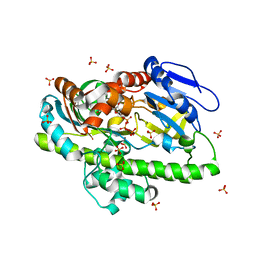 | | RNA ligase RtcB from Pyrococcus horikoshii in complex with Mn2+ and GTP | | Descriptor: | CHLORIDE ION, GUANOSINE-5'-TRIPHOSPHATE, MANGANESE (II) ION, ... | | Authors: | Jacewicz, A, Dantuluri, S, Shuman, S. | | Deposit date: | 2022-06-16 | | Release date: | 2022-10-12 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (2.47 Å) | | Cite: | Structures of RNA ligase RtcB in complexes with divalent cations and GTP.
Rna, 28, 2022
|
|
8DCG
 
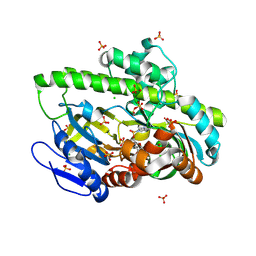 | | Structure of guanylylated RNA ligase RtcB from Pyrococcus horikoshii | | Descriptor: | CHLORIDE ION, GUANOSINE-5'-MONOPHOSPHATE, SULFATE ION, ... | | Authors: | Jacewicz, A, Dantuluri, S, Shuman, S. | | Deposit date: | 2022-06-16 | | Release date: | 2022-10-12 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (2.35 Å) | | Cite: | Structures of RNA ligase RtcB in complexes with divalent cations and GTP.
Rna, 28, 2022
|
|
8DCF
 
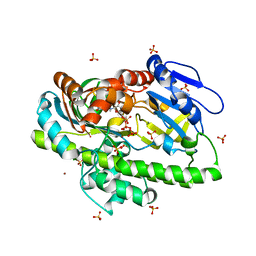 | | RNA ligase RtcB from Pyrococcus horikoshii in complex with Cu2+ and GTP | | Descriptor: | CHLORIDE ION, COPPER (II) ION, GLYCEROL, ... | | Authors: | Jacewicz, A, Dantuluri, S, Shuman, S. | | Deposit date: | 2022-06-16 | | Release date: | 2022-10-12 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (2.42 Å) | | Cite: | Structures of RNA ligase RtcB in complexes with divalent cations and GTP.
Rna, 28, 2022
|
|
8DCB
 
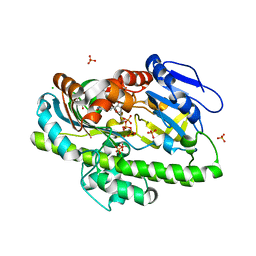 | | RNA ligase RtcB from Pyrococcus horikoshii in complex with Ni2+ and GTP | | Descriptor: | CHLORIDE ION, GLYCEROL, GUANOSINE-5'-TRIPHOSPHATE, ... | | Authors: | Jacewicz, A, Dantuluri, S, Shuman, S. | | Deposit date: | 2022-06-16 | | Release date: | 2022-10-12 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Structures of RNA ligase RtcB in complexes with divalent cations and GTP.
Rna, 28, 2022
|
|
8DCA
 
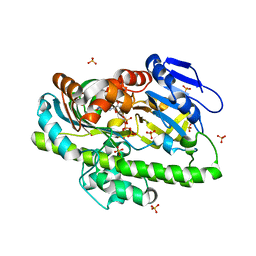 | | RNA ligase RtcB from Pyrococcus horikoshii in complex with Co2+ and GTP | | Descriptor: | COBALT (II) ION, GLYCEROL, GUANOSINE-5'-TRIPHOSPHATE, ... | | Authors: | Jacewicz, A, Dantuluri, S, Shuman, S. | | Deposit date: | 2022-06-16 | | Release date: | 2022-10-12 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (2.43 Å) | | Cite: | Structures of RNA ligase RtcB in complexes with divalent cations and GTP.
Rna, 28, 2022
|
|
1J5D
 
 | | SOLUTION STRUCTURE OF OXIDIZED PARAMAGNETIC CU(II) PLASTOCYANIN FROM SYNECHOCYSTIS PCC6803-MINIMIZED AVERAGE STRUCTURE | | Descriptor: | COPPER (II) ION, PLASTOCYANIN | | Authors: | Bertini, I, Ciurli, S, Dikiy, A, Fernandez, C.O, Luchinat, C, Safarov, N, Shumilin, S, Vila, A.J. | | Deposit date: | 2002-04-02 | | Release date: | 2002-04-10 | | Last modified: | 2023-12-27 | | Method: | SOLUTION NMR | | Cite: | The first solution structure of a paramagnetic copper(II) protein: the case of oxidized plastocyanin from the cyanobacterium Synechocystis PCC6803.
J.Am.Chem.Soc., 123, 2001
|
|
1J5C
 
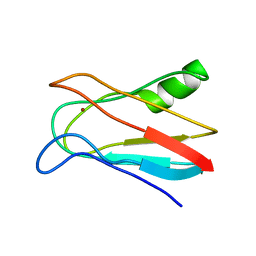 | | SOLUTION STRUCTURE OF OXIDIZED PARAMAGNETIC CU(II) PLASTOCYANIN FROM SYNECHOCYSTIS PCC6803 | | Descriptor: | COPPER (II) ION, PLASTOCYANIN | | Authors: | Bertini, I, Ciurli, S, Dikiy, A, Fernandez, C.O, Luchinat, C, Safarov, N, Shumilin, S, Vila, A.J. | | Deposit date: | 2002-04-02 | | Release date: | 2002-04-10 | | Last modified: | 2023-12-27 | | Method: | SOLUTION NMR | | Cite: | The first solution structure of a paramagnetic copper(II) protein: the case of oxidized plastocyanin from the cyanobacterium Synechocystis PCC6803.
J.Am.Chem.Soc., 123, 2001
|
|
6E33
 
 | | Crystal Structure of Pho7-DNA complex | | Descriptor: | DNA (5'-D(*GP*AP*TP*TP*TP*GP*AP*AP*TP*GP*TP*CP*CP*GP*AP*AP*GP*GP*AP*T)-3'), DNA (5'-D(*TP*CP*CP*TP*TP*CP*GP*GP*AP*CP*AP*TP*TP*CP*AP*AP*AP*TP*CP*A)-3'), Uncharacterized transcriptional regulatory protein C27B12.11c, ... | | Authors: | Garg, A, Goldgur, Y, Shuman, S. | | Deposit date: | 2018-07-13 | | Release date: | 2018-10-24 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (1.705 Å) | | Cite: | Distinctive structural basis for DNA recognition by the fission yeast Zn2Cys6 transcription factor Pho7 and its role in phosphate homeostasis.
Nucleic Acids Res., 46, 2018
|
|
7SJR
 
 | | Cryo-EM structure of AdnA-AdnB(W325A) in complex with DNA and AMPPNP | | Descriptor: | DNA (70-MER), DNA helicase, IRON/SULFUR CLUSTER, ... | | Authors: | Wang, J, Warren, G.M, Shuman, S, Patel, D.J. | | Deposit date: | 2021-10-18 | | Release date: | 2021-12-22 | | Last modified: | 2024-06-05 | | Method: | ELECTRON MICROSCOPY (3.8 Å) | | Cite: | Structure-activity relationships at a nucleobase-stacking tryptophan required for chemomechanical coupling in the DNA resecting motor-nuclease AdnAB.
Nucleic Acids Res., 50, 2022
|
|
7LFQ
 
 | | Pyrococcus RNA ligase | | Descriptor: | DNA (5'-D(*AP*TP*GP*TP*CP*C)-3'), POTASSIUM ION, RNA-splicing ligase RtcB, ... | | Authors: | Goldgur, Y, Shuman, S, Banerjee, A. | | Deposit date: | 2021-01-18 | | Release date: | 2021-03-03 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Structure of 3'-PO 4 /5'-OH RNA ligase RtcB in complex with a 5'-OH oligonucleotide.
Rna, 27, 2021
|
|
7LHL
 
 | |
7MI2
 
 | | Seb1-G476S RNA binding domain | | Descriptor: | DI(HYDROXYETHYL)ETHER, GLYCEROL, Rpb7-binding protein seb1 | | Authors: | Jacewicz, A, Shuman, S. | | Deposit date: | 2021-04-16 | | Release date: | 2021-07-14 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (1.4 Å) | | Cite: | Genetic screen for suppression of transcriptional interference identifies a gain-of-function mutation in Pol2 termination factor Seb1.
Proc.Natl.Acad.Sci.USA, 118, 2021
|
|
8TKZ
 
 | |
7U1Y
 
 | | Structure of SPAC806.04c protein from fission yeast bound to AlF4 and Co2+ | | Descriptor: | COBALT (II) ION, Damage-control phosphatase SPAC806.04c, POTASSIUM ION, ... | | Authors: | Jacewicz, A, Sanchez, A.M, Shuman, S. | | Deposit date: | 2022-02-22 | | Release date: | 2022-06-01 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (1.81 Å) | | Cite: | Fission yeast Duf89 and Duf8901 are cobalt/nickel-dependent phosphatase-pyrophosphatases that act via a covalent aspartyl-phosphate intermediate.
J.Biol.Chem., 298, 2022
|
|
7U1V
 
 | | Structure of SPAC806.04c protein from fission yeast covalently bound to BeF3 | | Descriptor: | Damage-control phosphatase SPAC806.04c, NICKEL (II) ION, PHOSPHATE ION, ... | | Authors: | Jacewicz, A, Sanchez, A.M, Shuman, S. | | Deposit date: | 2022-02-22 | | Release date: | 2022-06-01 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Fission yeast Duf89 and Duf8901 are cobalt/nickel-dependent phosphatase-pyrophosphatases that act via a covalent aspartyl-phosphate intermediate.
J.Biol.Chem., 298, 2022
|
|
7U1X
 
 | |
6PPU
 
 | | Cryo-EM structure of AdnAB-AMPPNP-DNA complex | | Descriptor: | ATP-dependent DNA helicase (UvrD/REP), DNA (29-MER), IRON/SULFUR CLUSTER, ... | | Authors: | Jia, N, Unciuleac, M, Shuman, S, Patel, D.J. | | Deposit date: | 2019-07-08 | | Release date: | 2019-11-20 | | Last modified: | 2024-03-20 | | Method: | ELECTRON MICROSCOPY (3.5 Å) | | Cite: | Structures and single-molecule analysis of bacterial motor nuclease AdnAB illuminate the mechanism of DNA double-strand break resection.
Proc.Natl.Acad.Sci.USA, 116, 2019
|
|
6N0T
 
 | | tRNA ligase | | Descriptor: | ADENOSINE-5'-TRIPHOSPHATE, MANGANESE (II) ION, SULFATE ION, ... | | Authors: | Banerjee, A, Goldgur, Y, Shuman, S. | | Deposit date: | 2018-11-07 | | Release date: | 2019-01-09 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (2.511 Å) | | Cite: | Structure and two-metal mechanism of fungal tRNA ligase.
Nucleic Acids Res., 47, 2019
|
|
4XBA
 
 | | Hnt3 | | Descriptor: | Aprataxin-like protein, GLYCEROL, GUANOSINE, ... | | Authors: | Jacewicz, A, Chauleau, M, Shuman, S. | | Deposit date: | 2014-12-16 | | Release date: | 2015-06-03 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | DNA3'pp5'G de-capping activity of aprataxin: effect of cap nucleoside analogs and structural basis for guanosine recognition.
Nucleic Acids Res., 43, 2015
|
|
5COV
 
 | |
5COT
 
 | |
5COU
 
 | |
4YKL
 
 | | Hnt3 in complex with DNA and guanosine | | Descriptor: | Aprataxin-like protein, CHLORIDE ION, DNA (5'-D(*GP*AP*AP*TP*CP*AP*TP*AP*AP*C)-3'), ... | | Authors: | Jacewicz, A, Chauleau, M, Shuman, S. | | Deposit date: | 2015-03-04 | | Release date: | 2015-06-03 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (2.25 Å) | | Cite: | DNA3'pp5'G de-capping activity of aprataxin: effect of cap nucleoside analogs and structural basis for guanosine recognition.
Nucleic Acids Res., 43, 2015
|
|
