6DAM
 
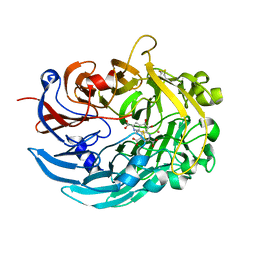 | | Crystal structure of lanthanide-dependent methanol dehydrogenase XoxF from Methylomicrobium buryatense 5G | | Descriptor: | LANTHANUM (III) ION, Lanthanide-dependent methanol dehydrogenase XoxF, PYRROLOQUINOLINE QUINONE, ... | | Authors: | Deng, Y, Ro, S.Y, Rosenzweig, A.C. | | Deposit date: | 2018-05-01 | | Release date: | 2018-09-05 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | Structure and function of the lanthanide-dependent methanol dehydrogenase XoxF from the methanotroph Methylomicrobium buryatense 5GB1C.
J. Biol. Inorg. Chem., 23, 2018
|
|
4M1H
 
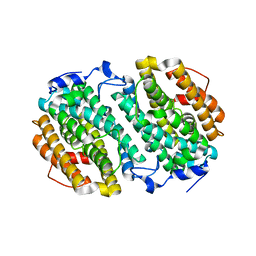 | | X-ray crystal structure of Chlamydia trachomatis apo NrdB | | Descriptor: | Ribonucleoside-diphosphate reductase subunit beta | | Authors: | Boal, A.K, Rosenzweig, A.C. | | Deposit date: | 2013-08-02 | | Release date: | 2013-08-21 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (1.695 Å) | | Cite: | Structural Basis for Assembly of the Mn(IV)/Fe(III) Cofactor in the Class Ic Ribonucleotide Reductase from Chlamydia trachomatis.
Biochemistry, 52, 2013
|
|
4M1F
 
 | |
3RGB
 
 | |
3RFA
 
 | | X-ray structure of RlmN from Escherichia coli in complex with S-adenosylmethionine | | Descriptor: | IRON/SULFUR CLUSTER, Ribosomal RNA large subunit methyltransferase N, S-ADENOSYLMETHIONINE | | Authors: | Boal, A.K, Grove, T.L, McLaughlin, M.I, Yennawar, N, Booker, S.J, Rosenzweig, A.C. | | Deposit date: | 2011-04-05 | | Release date: | 2011-05-11 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (2.05 Å) | | Cite: | Structural basis for methyl transfer by a radical SAM enzyme.
Science, 332, 2011
|
|
3RFR
 
 | |
3RF9
 
 | | X-ray structure of RlmN from Escherichia coli | | Descriptor: | (4R)-2-METHYLPENTANE-2,4-DIOL, IRON/SULFUR CLUSTER, Ribosomal RNA large subunit methyltransferase N | | Authors: | Boal, A.K, Grove, T.L, McLaughlin, M.I, Yennawar, N, Booker, S.J, Rosenzweig, A.C. | | Deposit date: | 2011-04-05 | | Release date: | 2011-05-11 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Structural basis for methyl transfer by a radical SAM enzyme.
Science, 332, 2011
|
|
3EWK
 
 | |
3RJ6
 
 | |
3RJN
 
 | |
3FDC
 
 | | Crystal Structure of Avidin | | Descriptor: | Avidin, iron(II) tetracyano-5-(2-Oxo-hexahydro-thieno[3,4-d]imidazol-6-yl)-pentanoic acid (4'-methyl-[2,2']bipyridinyl-4-ylmethyl)-amide | | Authors: | Barker, K.D, Sazinsky, M.H, Eckermann, A.L, Abajian, C, Hartings, M.R, Rosenzweig, A.C, Meade, T.J. | | Deposit date: | 2008-11-25 | | Release date: | 2009-12-01 | | Last modified: | 2024-10-09 | | Method: | X-RAY DIFFRACTION (3.1 Å) | | Cite: | Protein Binding and the Electronic Properties of Iron(II) Complexes: An Electrochemical and Optical Investigation of Outer Sphere Effects.
Bioconjug.Chem., 20, 2009
|
|
3FRY
 
 | | Crystal structure of the CopA C-terminal metal binding domain | | Descriptor: | CITRIC ACID, Probable copper-exporting P-type ATPase A | | Authors: | Agarwal, S, Sazinsky, M, Arguello, J, Rosenzweig, A.C. | | Deposit date: | 2009-01-08 | | Release date: | 2010-01-12 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Structure and interactions of the C-terminal metal binding domain of Archaeoglobus fulgidus CopA.
Proteins, 78, 2010
|
|
3G5W
 
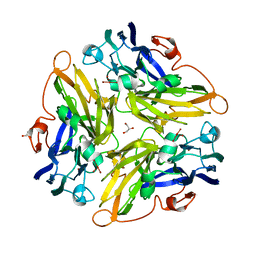 | | Crystal structure of Blue Copper Oxidase from Nitrosomonas europaea | | Descriptor: | COPPER (II) ION, CU-O LINKAGE, CU-O-CU LINKAGE, ... | | Authors: | Lawton, T.J, Sayavedra-Soto, L.A, Arp, D.J, Rosenzweig, A.C. | | Deposit date: | 2009-02-05 | | Release date: | 2009-02-17 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Crystal structure of a two-domain multicopper oxidase: implications for the evolution of multicopper blue proteins.
J.Biol.Chem., 284, 2009
|
|
4OJ8
 
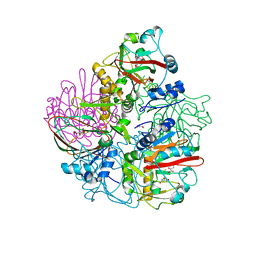 | | Crystal structure of carbapenem synthase in complex with (3S,5S)-carbapenam | | Descriptor: | (2S,5S)-7-oxo-1-azabicyclo[3.2.0]heptane-2-carboxylic acid, (5R)-carbapenem-3-carboxylate synthase, 2-OXOGLUTARIC ACID, ... | | Authors: | Boal, A.K, Rosenzweig, A.C. | | Deposit date: | 2014-01-20 | | Release date: | 2014-04-02 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Mechanism of the C5 stereoinversion reaction in the biosynthesis of carbapenem antibiotics.
Science, 343, 2014
|
|
4PL1
 
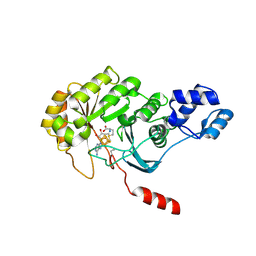 | |
4PL2
 
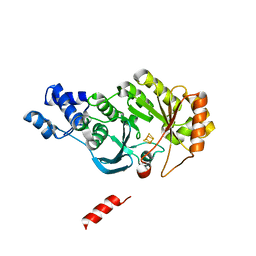 | |
4O65
 
 | | Crystal structure of the cupredoxin domain of amoB from Nitrosocaldus yellowstonii | | Descriptor: | COPPER (II) ION, Putative archaeal ammonia monooxygenase subunit B, SULFATE ION | | Authors: | Lawton, T.J, Ham, J, Sun, T, Rosenzweig, A.C. | | Deposit date: | 2013-12-20 | | Release date: | 2014-04-02 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (1.796 Å) | | Cite: | Structural conservation of the B subunit in the ammonia monooxygenase/particulate methane monooxygenase superfamily.
Proteins, 82, 2014
|
|
4NS2
 
 | |
4PHZ
 
 | |
4PI2
 
 | |
4N83
 
 | |
4PI0
 
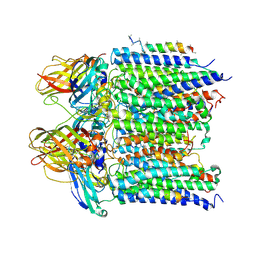 | |
7S4K
 
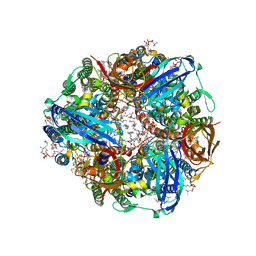 | | CryoEM structure of Methylococcus capsulatus (Bath) pMMO in a native lipid nanodisc at 2.34 Angstrom resolution | | Descriptor: | 1,2-DIDECANOYL-SN-GLYCERO-3-PHOSPHOCHOLINE, 1,2-dihexanoyl-sn-glycero-3-phosphocholine, Ammonia monooxygenase/methane monooxygenase, ... | | Authors: | Koo, C.W, Rosenzweig, A.C. | | Deposit date: | 2021-09-09 | | Release date: | 2022-03-30 | | Last modified: | 2024-06-05 | | Method: | ELECTRON MICROSCOPY (2.36 Å) | | Cite: | Recovery of particulate methane monooxygenase structure and activity in a lipid bilayer.
Science, 375, 2022
|
|
7S4J
 
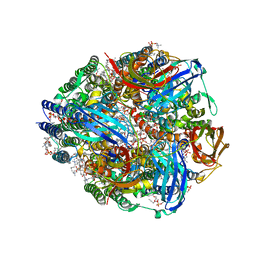 | | CryoEM structure of Methylococcus capsulatus (Bath) pMMO in a native lipid nanodisc at 2.16 Angstrom resolution | | Descriptor: | 1,2-DIDECANOYL-SN-GLYCERO-3-PHOSPHOCHOLINE, 1,2-dihexanoyl-sn-glycero-3-phosphocholine, Ammonia monooxygenase/methane monooxygenase, ... | | Authors: | Koo, C.W, Rosenzweig, A.C. | | Deposit date: | 2021-09-09 | | Release date: | 2022-03-30 | | Last modified: | 2024-06-05 | | Method: | ELECTRON MICROSCOPY (2.16 Å) | | Cite: | Recovery of particulate methane monooxygenase structure and activity in a lipid bilayer.
Science, 375, 2022
|
|
7S4H
 
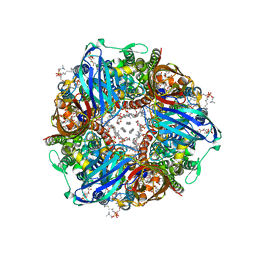 | | CryoEM structure of Methylococcus capsulatus (Bath) pMMO in a native lipid nanodisc at 2.14 Angstrom resolution | | Descriptor: | 1,2-DIDECANOYL-SN-GLYCERO-3-PHOSPHOCHOLINE, 1,2-dihexanoyl-sn-glycero-3-phosphocholine, Ammonia monooxygenase/methane monooxygenase, ... | | Authors: | Koo, C.W, Rosenzweig, A.C. | | Deposit date: | 2021-09-08 | | Release date: | 2022-03-30 | | Last modified: | 2024-06-05 | | Method: | ELECTRON MICROSCOPY (2.14 Å) | | Cite: | Recovery of particulate methane monooxygenase structure and activity in a lipid bilayer.
Science, 375, 2022
|
|
