6I40
 
 | | Crystal structure of murine neuroglobin bound to CO at 15K under illumination using optical fiber | | Descriptor: | ACETATE ION, CARBON MONOXIDE, FORMIC ACID, ... | | Authors: | Savino, C, Montemiglio, L.C, Ardiccioni, C, Exertier, C, Vallone, B. | | Deposit date: | 2018-11-08 | | Release date: | 2019-09-11 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Ligand pathways in neuroglobin revealed by low-temperature photodissociation and docking experiments.
Iucrj, 6, 2019
|
|
8A58
 
 | |
1HGZ
 
 | | Filamentous Bacteriophage PH75 | | Descriptor: | PH75 INOVIRUS MAJOR COAT PROTEIN | | Authors: | Pederson, D.M, Welsh, L.C, Marvin, D.A, Sampson, M, Perham, R.N, Yu, M, Slater, M.R. | | Deposit date: | 2000-12-17 | | Release date: | 2001-06-01 | | Last modified: | 2024-02-14 | | Method: | FIBER DIFFRACTION (2.4 Å) | | Cite: | The Protein Capsid of Filamentous Bacteriophage Ph75 from Thermus Thermophilus
J.Mol.Biol., 309, 2001
|
|
1HH0
 
 | | Filamentous Bacteriophage PH75 | | Descriptor: | PH75 INOVIRUS MAJOR COAT PROTEIN | | Authors: | Pederson, D.M, Welsh, L.C, Marvin, D.A, Sampson, M, Perham, R.N, Yu, M, Slater, M.R. | | Deposit date: | 2000-12-17 | | Release date: | 2001-06-01 | | Last modified: | 2024-02-14 | | Method: | FIBER DIFFRACTION (2.4 Å) | | Cite: | The Protein Capsid of Filamentous Bacteriophage Ph75 from Thermus Thermophilus
J.Mol.Biol., 309, 2001
|
|
1HGV
 
 | | Filamentous Bacteriophage PH75 | | Descriptor: | PH75 INOVIRUS MAJOR COAT PROTEIN | | Authors: | Pederson, D.M, Welsh, L.C, Marvin, D.A, Sampson, M, Perham, R.N, Yu, M, Slater, M.R. | | Deposit date: | 2000-12-15 | | Release date: | 2001-06-01 | | Last modified: | 2023-12-13 | | Method: | FIBER DIFFRACTION (2.4 Å) | | Cite: | The Protein Capsid of Filamentous Bacteriophage Ph75 from Thermus Thermophilus
J.Mol.Biol., 309, 2001
|
|
4LZT
 
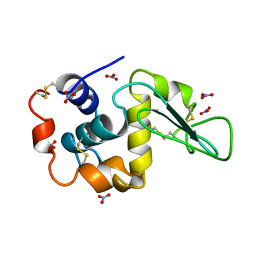 | | ATOMIC RESOLUTION REFINEMENT OF TRICLINIC HEW LYSOZYME AT 295K | | Descriptor: | LYSOZYME, NITRATE ION | | Authors: | Walsh, M.A, Schneider, T, Sieker, L.C, Dauter, Z, Lamzin, V, Wilson, K.S. | | Deposit date: | 1997-03-31 | | Release date: | 1998-04-01 | | Last modified: | 2023-08-09 | | Method: | X-RAY DIFFRACTION (0.95 Å) | | Cite: | Refinement of triclinic hen egg-white lysozyme at atomic resolution.
Acta Crystallogr.,Sect.D, 54, 1998
|
|
6S53
 
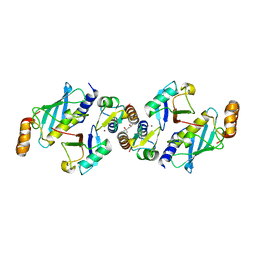 | | Crystal structure of TRIM21 RING domain in complex with an isopeptide-linked Ube2N~ubiquitin conjugate | | Descriptor: | (4S)-2-METHYL-2,4-PENTANEDIOL, E3 ubiquitin-protein ligase TRIM21, Polyubiquitin-C, ... | | Authors: | Kiss, L, Boland, A, Neuhaus, D, James, L.C. | | Deposit date: | 2019-06-30 | | Release date: | 2019-09-11 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | A tri-ionic anchor mechanism drives Ube2N-specific recruitment and K63-chain ubiquitination in TRIM ligases.
Nat Commun, 10, 2019
|
|
6RSS
 
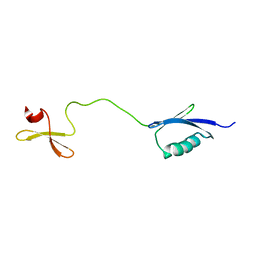 | | Solution structure of the fourth WW domain of WWP2 with GB1-tag | | Descriptor: | NEDD4-like E3 ubiquitin-protein ligase WWP2 | | Authors: | Wahl, L.C, Watt, J.E, Tolchard, J, Blumenschein, T.M.A, Chantry, A. | | Deposit date: | 2019-05-22 | | Release date: | 2019-10-09 | | Last modified: | 2024-06-19 | | Method: | SOLUTION NMR | | Cite: | Smad7 Binds Differently to Individual and Tandem WW3 and WW4 Domains of WWP2 Ubiquitin Ligase Isoforms.
Int J Mol Sci, 20, 2019
|
|
7SZI
 
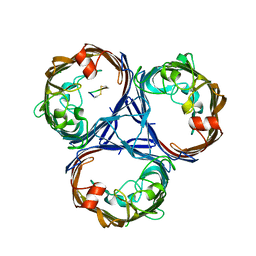 | | Cryo-EM structure of OmpK36-TraN mating pair stabilization proteins from carbapenem-resistant Klebsiella pneumoniae | | Descriptor: | OmpK36, TraN | | Authors: | Beltran, L.C, Seddon, C, Beis, K, Frankel, G, Egelman, E.H. | | Deposit date: | 2021-11-27 | | Release date: | 2022-06-08 | | Last modified: | 2022-07-13 | | Method: | ELECTRON MICROSCOPY (2.7 Å) | | Cite: | Mating pair stabilization mediates bacterial conjugation species specificity.
Nat Microbiol, 7, 2022
|
|
6TYW
 
 | |
6TYZ
 
 | |
6TYV
 
 | |
6TYX
 
 | |
6TYU
 
 | |
6U7D
 
 | |
6TYT
 
 | |
1LZT
 
 | | REFINEMENT OF TRICLINIC LYSOZYME | | Descriptor: | HEN EGG WHITE LYSOZYME | | Authors: | Hodsdon, J.M, Brown, G.M, Sieker, L.C, Jensen, L.H. | | Deposit date: | 1985-04-01 | | Release date: | 1985-07-18 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (1.97 Å) | | Cite: | Refinement of triclinic lysozyme: I. Fourier and least-squares methods.
Acta Crystallogr.,Sect.B, 46, 1990
|
|
4GBI
 
 | | Crystal structure of aspart insulin at pH 6.5 | | Descriptor: | CHLORIDE ION, Insulin A chain, Insulin B chain, ... | | Authors: | Lima, L.M.T.R, Favero-Retto, M.P, Palmieri, L.C. | | Deposit date: | 2012-07-27 | | Release date: | 2013-06-12 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (2.502 Å) | | Cite: | A T3R3 hexamer of the human insulin variant B28Asp.
Biophys.Chem., 173, 2013
|
|
4GBL
 
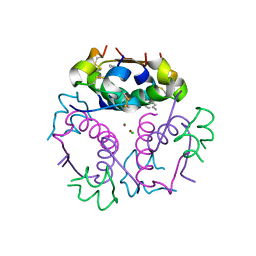 | | Crystal structure of aspart insulin at pH 8.5 | | Descriptor: | CHLORIDE ION, Insulin A chain, Insulin B chain, ... | | Authors: | Lima, L.M.T.R, Favero-Retto, M.P, Palmieri, L.C. | | Deposit date: | 2012-07-27 | | Release date: | 2013-06-12 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | A T3R3 hexamer of the human insulin variant B28Asp.
Biophys.Chem., 173, 2013
|
|
1RV7
 
 | | Crystal structures of a Multidrug-Resistant HIV-1 Protease Reveal an Expanded Active Site Cavity | | Descriptor: | N-{1-BENZYL-4-[2-(2,6-DIMETHYL-PHENOXY)-ACETYLAMINO]-3-HYDROXY-5-PHENYL-PENTYL}-3-METHYL-2-(2-OXO-TETRAHYDRO-PYRIMIDIN-1-YL)-BUTYRAMIDE, protease | | Authors: | Logsdon, B.C, Vickrey, J.F, Martin, P, Proteasa, G, Koepke, J.I, Terlecky, S.R, Wawrzak, Z, Winters, M.A, Merigan, T.C, Kovari, L.C. | | Deposit date: | 2003-12-12 | | Release date: | 2004-12-14 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Crystal structures of a multidrug-resistant human immunodeficiency virus type 1 protease reveal an expanded active-site cavity.
J.Virol., 78, 2004
|
|
1RPI
 
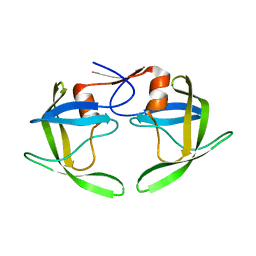 | | Crystal structures of a Multidrug-Resistant HIV-1 Protease Reveal an Expanded Active Site Cavity | | Descriptor: | alpha-D-glucopyranose, protease | | Authors: | Logsdon, B.C, Vickrey, J.F, Martin, P, Proteasa, G, Koepke, J.I, Terlecky, S.R, Wawrzak, Z, Winters, M.A, Merigan, T.C, Kovari, L.C. | | Deposit date: | 2003-12-03 | | Release date: | 2004-12-07 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (1.86 Å) | | Cite: | Crystal structures of a multidrug-resistant human immunodeficiency virus type 1 protease reveal an expanded active-site cavity.
J.Virol., 78, 2004
|
|
1UBV
 
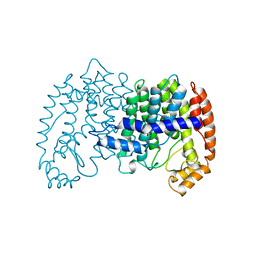 | |
1UBX
 
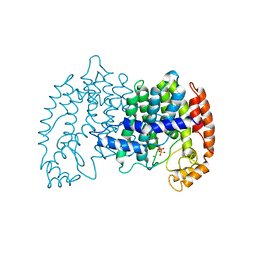 | | STRUCTURE OF FARNESYL PYROPHOSPHATE SYNTHETASE | | Descriptor: | FARNESYL DIPHOSPHATE, FARNESYL DIPHOSPHATE SYNTHASE, MAGNESIUM ION | | Authors: | Tarshis, L.C, Proteau, P, Poulter, C.D, Sacchettini, J.C. | | Deposit date: | 1996-10-14 | | Release date: | 1997-03-12 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Regulation of product chain length by isoprenyl diphosphate synthases.
Proc.Natl.Acad.Sci.USA, 93, 1996
|
|
1UBW
 
 | | STRUCTURE OF FARNESYL PYROPHOSPHATE SYNTHETASE | | Descriptor: | FARNESYL DIPHOSPHATE SYNTHASE, GERANYL DIPHOSPHATE, MAGNESIUM ION | | Authors: | Tarshis, L.C, Proteau, P, Poulter, C.D, Sacchettini, J.C. | | Deposit date: | 1996-10-14 | | Release date: | 1997-03-12 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Regulation of product chain length by isoprenyl diphosphate synthases.
Proc.Natl.Acad.Sci.USA, 93, 1996
|
|
1UBY
 
 | | STRUCTURE OF FARNESYL PYROPHOSPHATE SYNTHETASE | | Descriptor: | DIMETHYLALLYL DIPHOSPHATE, FARNESYL DIPHOSPHATE SYNTHASE, MAGNESIUM ION | | Authors: | Tarshis, L.C, Proteau, P, Poulter, C.D, Sacchettini, J.C. | | Deposit date: | 1996-10-14 | | Release date: | 1997-03-12 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Regulation of product chain length by isoprenyl diphosphate synthases.
Proc.Natl.Acad.Sci.USA, 93, 1996
|
|
