7UHH
 
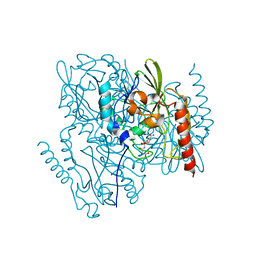 | | Time-Resolved Structure of Metallo Beta-Lactamase L1 in a Complex with Non-Hydrolyzed Moxalactam (20 ms snapshot) | | Descriptor: | (1R,6R,7R)-7-[(2R)-2-carboxypropanamido]-7-methoxy-3-methyl-8-oxo-5-oxa-1-azabicyclo[4.2.0]oct-2-ene-2-carboxylic acid, Putative metallo-beta-lactamase l1 (Beta-lactamase type ii) (Ec 3.5.2.6) (Penicillinase), ZINC ION | | Authors: | Wilamowski, M, Kim, Y, Sherrell, D.A, Lavens, A, Henning, R, Maltseva, N, Endres, M, Babnigg, G, Srajer, V, Joachimiak, A, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2022-03-27 | | Release date: | 2022-04-13 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Time-resolved beta-lactam cleavage by L1 metallo-beta-lactamase.
Nat Commun, 13, 2022
|
|
7UHP
 
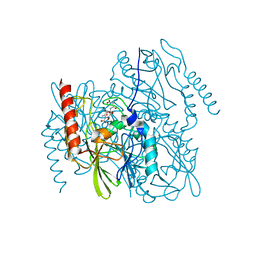 | | Time-Resolved Structure of Metallo Beta-Lactamase L1 in a Complex with Cleaved Moxalactam (2000 ms Snapshot) | | Descriptor: | (2R)-2-[(R)-carboxy{[(2R)-2-carboxy-2-(4-hydroxyphenyl)acetyl]amino}methoxymethyl]-5-{[(1-methyl-1H-tetrazol-5-yl)sulfanyl]methyl}-3,6-dihydro-2H-1,3-oxazine-4-carboxylic acid, Putative metallo-beta-lactamase l1 (Beta-lactamase type ii) (Ec 3.5.2.6) (Penicillinase), ZINC ION | | Authors: | Wilamowski, M, Kim, Y, Sherrell, D.A, Lavens, A, Henning, R, Maltseva, N, Endres, M, Babnigg, G, Srajer, V, Joachimiak, A, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2022-03-27 | | Release date: | 2022-04-13 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Time-resolved beta-lactam cleavage by L1 metallo-beta-lactamase.
Nat Commun, 13, 2022
|
|
7UHT
 
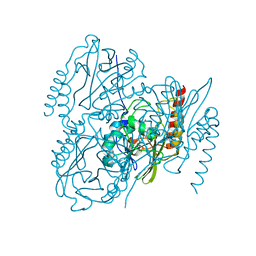 | | SSX Structure of Metallo Beta-Lactamase L1 with One Zinc in the Active Site | | Descriptor: | Putative metallo-beta-lactamase l1 (Beta-lactamase type ii) (Ec 3.5.2.6) (Penicillinase), ZINC ION | | Authors: | Wilamowski, M, Kim, Y, Sherrell, D.A, Lavens, A, Henning, R, Maltseva, N, Endres, M, Babnigg, G, Srajer, V, Joachimiak, A, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2022-03-27 | | Release date: | 2022-04-13 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Time-resolved beta-lactam cleavage by L1 metallo-beta-lactamase.
Nat Commun, 13, 2022
|
|
7UHK
 
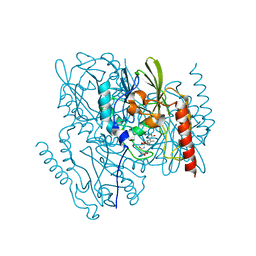 | | Time-Resolved Structure of Metallo Beta-Lactamase L1 in a Complex with Non-Hydrolyzed Moxalactam (80 ms Snapshot) | | Descriptor: | (1R,6R,7R)-7-[(2R)-2-carboxypropanamido]-7-methoxy-3-methyl-8-oxo-5-oxa-1-azabicyclo[4.2.0]oct-2-ene-2-carboxylic acid, Putative metallo-beta-lactamase l1 (Beta-lactamase type ii) (Ec 3.5.2.6) (Penicillinase), ZINC ION | | Authors: | Wilamowski, M, Kim, Y, Sherrell, D.A, Lavens, A, Henning, R, Maltseva, N, Endres, M, Babnigg, G, Srajer, V, Joachimiak, A, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2022-03-27 | | Release date: | 2022-04-13 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Time-resolved beta-lactam cleavage by L1 metallo-beta-lactamase.
Nat Commun, 13, 2022
|
|
7UHQ
 
 | | Time-Resolved Structure of Metallo Beta-Lactamase L1 in a Complex with Cleaved Moxalactam (4000 ms Snapshot) | | Descriptor: | (2R)-2-[(R)-carboxy{[(2R)-2-carboxy-2-(4-hydroxyphenyl)acetyl]amino}methoxymethyl]-5-{[(1-methyl-1H-tetrazol-5-yl)sulfanyl]methyl}-3,6-dihydro-2H-1,3-oxazine-4-carboxylic acid, Putative metallo-beta-lactamase l1 (Beta-lactamase type ii) (Ec 3.5.2.6) (Penicillinase), ZINC ION | | Authors: | Wilamowski, M, Kim, Y, Sherrell, D.A, Lavens, A, Henning, R, Maltseva, N, Endres, M, Babnigg, G, Srajer, V, Joachimiak, A, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2022-03-27 | | Release date: | 2022-04-13 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Time-resolved beta-lactam cleavage by L1 metallo-beta-lactamase.
Nat Commun, 13, 2022
|
|
7UHI
 
 | | Time-Resolved Structure of Metallo Beta-Lactamase L1 in a Complex with Non-Hydrolyzed Moxalactam (40 ms Snapshot) | | Descriptor: | (1R,6R,7R)-7-[(2R)-2-carboxypropanamido]-7-methoxy-3-methyl-8-oxo-5-oxa-1-azabicyclo[4.2.0]oct-2-ene-2-carboxylic acid, Putative metallo-beta-lactamase l1 (Beta-lactamase type ii) (Ec 3.5.2.6) (Penicillinase), ZINC ION | | Authors: | Wilamowski, M, Kim, Y, Sherrell, D.A, Lavens, A, Henning, R, Maltseva, N, Endres, M, Babnigg, G, Srajer, V, Joachimiak, A, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2022-03-27 | | Release date: | 2022-04-13 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Time-resolved beta-lactam cleavage by L1 metallo-beta-lactamase.
Nat Commun, 13, 2022
|
|
7UHL
 
 | | Time-Resolved Structure of Metallo Beta-Lactamase L1 in a Complex with Non-Hydrolyzed Moxalactam (100 ms Snapshot) | | Descriptor: | (1R,6R,7R)-7-[(2R)-2-carboxypropanamido]-7-methoxy-3-methyl-8-oxo-5-oxa-1-azabicyclo[4.2.0]oct-2-ene-2-carboxylic acid, Putative metallo-beta-lactamase l1 (Beta-lactamase type ii) (Ec 3.5.2.6) (Penicillinase), ZINC ION | | Authors: | Wilamowski, M, Kim, Y, Sherrell, D.A, Lavens, A, Henning, R, Maltseva, N, Endres, M, Babnigg, G, Srajer, V, Joachimiak, A, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2022-03-27 | | Release date: | 2022-04-13 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Time-resolved beta-lactam cleavage by L1 metallo-beta-lactamase.
Nat Commun, 13, 2022
|
|
7UHR
 
 | | Time-Resolved Structure of Metallo Beta-Lactamase L1 Before Reaction (Dark-Set) | | Descriptor: | Putative metallo-beta-lactamase l1 (Beta-lactamase type ii) (Ec 3.5.2.6) (Penicillinase), ZINC ION | | Authors: | Wilamowski, M, Kim, Y, Sherrell, D.A, Lavens, A, Henning, R, Maltseva, N, Endres, M, Babnigg, G, Srajer, V, Joachimiak, A, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2022-03-27 | | Release date: | 2022-04-13 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Time-resolved beta-lactam cleavage by L1 metallo-beta-lactamase.
Nat Commun, 13, 2022
|
|
7UHM
 
 | | Time-Resolved Structure of Metallo Beta-Lactamase L1 in a Complex with Cleaved Moxalactam (150 ms Snapshot) | | Descriptor: | (2R)-2-[(R)-carboxy{[(2R)-2-carboxy-2-(4-hydroxyphenyl)acetyl]amino}methoxymethyl]-5-{[(1-methyl-1H-tetrazol-5-yl)sulfanyl]methyl}-3,6-dihydro-2H-1,3-oxazine-4-carboxylic acid, Putative metallo-beta-lactamase l1 (Beta-lactamase type ii) (Ec 3.5.2.6) (Penicillinase), ZINC ION | | Authors: | Wilamowski, M, Kim, Y, Sherrell, D.A, Lavens, A, Henning, R, Maltseva, N, Endres, M, Babnigg, G, Srajer, V, Joachimiak, A, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2022-03-27 | | Release date: | 2022-06-29 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Time-resolved beta-lactam cleavage by L1 metallo-beta-lactamase.
Nat Commun, 13, 2022
|
|
7UHO
 
 | | Time-Resolved Structure of Metallo Beta-Lactamase L1 in a Complex with Cleaved Moxalactam (500 ms Snapshot) | | Descriptor: | (2R)-2-[(R)-carboxy{[(2R)-2-carboxy-2-(4-hydroxyphenyl)acetyl]amino}methoxymethyl]-5-{[(1-methyl-1H-tetrazol-5-yl)sulfanyl]methyl}-3,6-dihydro-2H-1,3-oxazine-4-carboxylic acid, Putative metallo-beta-lactamase l1 (Beta-lactamase type ii) (Ec 3.5.2.6) (Penicillinase), ZINC ION | | Authors: | Wilamowski, M, Kim, Y, Sherrell, D.A, Lavens, A, Henning, R, Maltseva, N, Endres, M, Babnigg, G, Srajer, V, Joachimiak, A, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2022-03-27 | | Release date: | 2022-07-13 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Time-resolved beta-lactam cleavage by L1 metallo-beta-lactamase.
Nat Commun, 13, 2022
|
|
4ZTK
 
 | | Transpeptidase domain of FtsI4 D,D-transpeptidase from Legionella pneumophila. | | Descriptor: | 2-[N-CYCLOHEXYLAMINO]ETHANE SULFONIC ACID, Cell division protein FtsI/penicillin binding protein 2 | | Authors: | CUFF, M, OSIPIUK, J, WU, R, ENDRES, M, JOACHIMIAK, A, Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2015-05-14 | | Release date: | 2015-05-27 | | Last modified: | 2019-12-25 | | Method: | X-RAY DIFFRACTION (2.104 Å) | | Cite: | Transpeptidase domain of FtsI4 D,D-transpeptidase from Legionella pneumophila.
to be published
|
|
5BP8
 
 | | ent-Copalyl diphosphate synthase from Streptomyces platensis | | Descriptor: | 1,2-ETHANEDIOL, Ent-copalyl diphosphate synthase, SULFATE ION | | Authors: | Osipiuk, J, Hatzos-Skintges, C, Endres, M, Babnigg, G, Rudolf, J.D, Ma, M, Chang, C.-Y, Shen, B, Joachimiak, A, Enzyme Discovery for Natural Product Biosynthesis (NatPro), Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2015-05-27 | | Release date: | 2015-06-10 | | Last modified: | 2019-12-25 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Structure of the ent-Copalyl Diphosphate Synthase PtmT2 from Streptomyces platensis CB00739, a Bacterial Type II Diterpene Synthase.
J.Am.Chem.Soc., 138, 2016
|
|
5C0P
 
 | | The crystal structure of endo-arabinase from Bacteroides thetaiotaomicron VPI-5482 | | Descriptor: | 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, CHLORIDE ION, Endo-arabinase, ... | | Authors: | Tan, K, Cuff, M, Joachimiak, G, Endres, M, Joachimiak, A, Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2015-06-12 | | Release date: | 2015-07-01 | | Last modified: | 2019-12-25 | | Method: | X-RAY DIFFRACTION (1.532 Å) | | Cite: | The crystal structure of endo-arabinase from Bacteroides thetaiotaomicron VPI-5482
To Be Published
|
|
5CR9
 
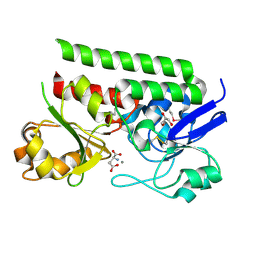 | | Crystal structure of ABC-type Fe3+-hydroxamate transport system from Saccharomonospora viridis DSM 43017 | | Descriptor: | ABC-type Fe3+-hydroxamate transport system, periplasmic component, GLUTAMIC ACID, ... | | Authors: | Nocek, B, Cuff, M, Mack, J, Endres, M, Joachimiak, A, Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2015-07-22 | | Release date: | 2015-08-12 | | Last modified: | 2019-12-25 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Crystal structure of ABC-type Fe3+-hydroxamate transport system from Saccharomonospora viridis DSM 43017
To Be Published
|
|
6VWW
 
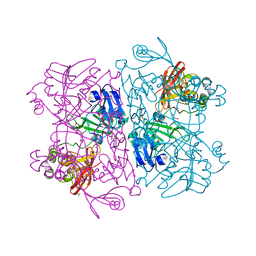 | | Crystal Structure of NSP15 Endoribonuclease from SARS CoV-2. | | Descriptor: | ACETIC ACID, CHLORIDE ION, GLYCEROL, ... | | Authors: | Kim, Y, Jedrzejczak, R, Maltseva, N, Endres, M, Godzik, A, Michalska, K, Joachimiak, A, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2020-02-20 | | Release date: | 2020-03-04 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Crystal structure of Nsp15 endoribonuclease NendoU from SARS-CoV-2.
Protein Sci., 29, 2020
|
|
5TF0
 
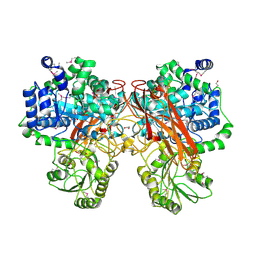 | | Crystal Structure of Glycosil Hydrolase Family 3 N-Terminal Domain Protein from Bacteroides intestinalis | | Descriptor: | 1,2-ETHANEDIOL, Glycosyl hydrolase family 3 N-terminal domain protein, MAGNESIUM ION | | Authors: | Kim, Y, Hatzos-Skintges, C, Endres, M, Babnigg, G, Joachimiak, A, MCSG, Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2016-09-23 | | Release date: | 2016-10-05 | | Last modified: | 2019-12-25 | | Method: | X-RAY DIFFRACTION (2.2021 Å) | | Cite: | Crystal Structure of Glycosil Hydrolase Family 3 N-Terminal Domain Protein from Bacteroides intestinalis
To Be Published, 2016
|
|
5TF3
 
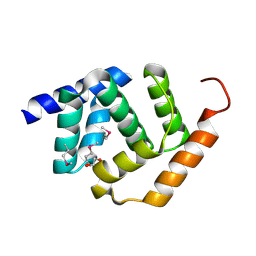 | | Crystal Structure of Protein of Unknown Function YPO2564 from Yersinia pestis | | Descriptor: | 1,2-ETHANEDIOL, Putative membrane protein | | Authors: | Kim, Y, Chhor, G, Endres, M, Babnigg, G, Anderson, W.F, Crosson, S, Joachimiak, A, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2016-09-23 | | Release date: | 2016-10-19 | | Last modified: | 2018-01-24 | | Method: | X-RAY DIFFRACTION (2.001 Å) | | Cite: | Crystal Structure of Protein of Unknown Function YPO2564 from Yersinia pestis
To Be Published
|
|
5TTX
 
 | |
5UFH
 
 | | The crystal structure of a LacI-type transcription regulator from Bifidobacterium animalis subsp. lactis DSM 10140 | | Descriptor: | GLYCEROL, LacI-type transcriptional regulator, NITRATE ION | | Authors: | Tan, K, Li, H, Endres, M, Joachimiak, A, Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2017-01-04 | | Release date: | 2017-01-18 | | Last modified: | 2020-01-01 | | Method: | X-RAY DIFFRACTION (1.45 Å) | | Cite: | The crystal structure of a LacI-type transcription regulator from Bifidobacterium animalis subsp. lactis DSM 10140
To Be Published
|
|
5UHJ
 
 | | The crystal structure of a natural product biosynthetic enzyme from Streptomyces sp. CB03234 | | Descriptor: | FORMIC ACID, Glyoxalase/bleomycin resisance protein/dioxygenase | | Authors: | Tan, K, Li, H, Endres, M, Phillips Jr, G.N, Joachimiak, A, Midwest Center for Structural Genomics (MCSG), Enzyme Discovery for Natural Product Biosynthesis (NatPro) | | Deposit date: | 2017-01-11 | | Release date: | 2017-01-25 | | Last modified: | 2020-09-23 | | Method: | X-RAY DIFFRACTION (1.75 Å) | | Cite: | The crystal structure of a natural product biosynthetic enzyme from Streptomyces sp. CB03234
To Be Published
|
|
5UJP
 
 | | The crystal structure of a glyoxalase/bleomycin resistance protein from Streptomyces sp. CB03234 | | Descriptor: | 2-(N-MORPHOLINO)-ETHANESULFONIC ACID, CALCIUM ION, Glyoxalase/bleomycin resisance protein/dioxygenase | | Authors: | Tan, K, Li, H, Endres, M, Phillips Jr, G.N, Joachimiak, A, Midwest Center for Structural Genomics (MCSG), Enzyme Discovery for Natural Product Biosynthesis (NatPro) | | Deposit date: | 2017-01-18 | | Release date: | 2017-02-22 | | Last modified: | 2020-01-01 | | Method: | X-RAY DIFFRACTION (1.42 Å) | | Cite: | The crystal structure of a glyoxalase/bleomycin resistance protein from Streptomyces sp. CB03234
To Be Published
|
|
5V4D
 
 | | Crystal Structure of the Protein of Unknown Function of the Conserved Rid Protein Family YyfA from Yersinia pestis | | Descriptor: | ACETIC ACID, CALCIUM ION, GLYCEROL, ... | | Authors: | Kim, Y, Chhor, G, Endres, M, Krishnan, A, Babnigg, G, Schneewind, O, Anderson, W.F, Joachimiak, A, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2017-03-09 | | Release date: | 2017-04-05 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Crystal Structure of the Protein of Unknown Function of the Conserved Rid Protein Family YyfA from Yersinia pestis
To Be Published
|
|
5UNC
 
 | | The crystal structure of PHOSPHOENOLPYRUVATE PHOSPHOMUTASE from Streptomyces platensis subsp. rosaceus | | Descriptor: | FORMIC ACID, L(+)-TARTARIC ACID, PHOSPHOENOLPYRUVATE PHOSPHOMUTASE, ... | | Authors: | Tan, K, Hatzos-Skintges, C, Endres, M, Phillips Jr, G.N, Joachimiak, A, Midwest Center for Structural Genomics (MCSG), Enzyme Discovery for Natural Product Biosynthesis (NatPro) | | Deposit date: | 2017-01-30 | | Release date: | 2017-02-22 | | Last modified: | 2020-07-29 | | Method: | X-RAY DIFFRACTION (1.71 Å) | | Cite: | The crystal structure of PHOSPHOENOLPYRUVATE PHOSPHOMUTASE from Streptomyces platensis subsp. rosaceus
To Be Published
|
|
5V4F
 
 | | Crystal Structure of the Protein of Unknown Function of the Conserved Rid Protein Family YyfB from Yersinia pestis | | Descriptor: | GLYCEROL, Putative translational inhibitor protein | | Authors: | Kim, Y, Chhor, G, Endres, M, Krishnan, A, Babnigg, G, Schneewind, O, Anderson, W.F, Joachimiak, A, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2017-03-09 | | Release date: | 2017-04-05 | | Last modified: | 2018-01-24 | | Method: | X-RAY DIFFRACTION (3.001 Å) | | Cite: | Crystal Structure of the Protein of Unknown Function of the Conserved Rid Protein Family YyfB from Yersinia pestis
To Be Published
|
|
5UVE
 
 | | Crystal Structure of the ABC Transporter Substrate-binding protein BAB1_0226 from Brucella abortus | | Descriptor: | CALCIUM ION, GLYCEROL, Substrate-binding region of ABC-type glycine betaine transport system | | Authors: | Kim, Y, Chhor, G, Endres, M, Hero, J, Babnigg, G, Crosson, S, Joachimiak, A, Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2017-02-20 | | Release date: | 2017-03-08 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Crystal Structure of Beta-barrel-like Protein of Unknown Function
To Be Published
|
|
