1G31
 
 | | GP31 CO-CHAPERONIN FROM BACTERIOPHAGE T4 | | Descriptor: | GP31, PHOSPHATE ION, POTASSIUM ION | | Authors: | Hunt, J.F, Van Der Vies, S.M, Henry, L, Deisenhofer, J. | | Deposit date: | 1998-03-27 | | Release date: | 1998-08-26 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Structural adaptations in the specialized bacteriophage T4 co-chaperonin Gp31 expand the size of the Anfinsen cage.
Cell(Cambridge,Mass.), 90, 1997
|
|
7PRC
 
 | | PHOTOSYNTHETIC REACTION CENTER FROM RHODOPSEUDOMONAS VIRIDIS (DG-420315 (TRIAZINE) COMPLEX) | | Descriptor: | 15-cis-1,2-dihydroneurosporene, 2-CHLORO-4-ETHYLAMINO-6-(R(+)-2'-CYANO-4-BUTYLAMINO)-1,3,5-TRIAZINE, BACTERIOCHLOROPHYLL B, ... | | Authors: | Lancaster, C.R.D, Michel, H. | | Deposit date: | 1997-08-01 | | Release date: | 1999-04-06 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (2.65 Å) | | Cite: | Refined crystal structures of reaction centres from Rhodopseudomonas viridis in complexes with the herbicide atrazine and two chiral atrazine derivatives also lead to a new model of the bound carotenoid.
J.Mol.Biol., 286, 1999
|
|
2PRC
 
 | | PHOTOSYNTHETIC REACTION CENTER FROM RHODOPSEUDOMONAS VIRIDIS (UBIQUINONE-2 COMPLEX) | | Descriptor: | 15-cis-1,2-dihydroneurosporene, BACTERIOCHLOROPHYLL B, BACTERIOPHEOPHYTIN B, ... | | Authors: | Lancaster, C.R.D, Michel, H. | | Deposit date: | 1997-07-29 | | Release date: | 1998-11-11 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (2.45 Å) | | Cite: | The coupling of light-induced electron transfer and proton uptake as derived from crystal structures of reaction centres from Rhodopseudomonas viridis modified at the binding site of the secondary quinone, QB.
Structure, 5, 1997
|
|
6PRC
 
 | | PHOTOSYNTHETIC REACTION CENTER FROM RHODOPSEUDOMONAS VIRIDIS (DG-420314 (TRIAZINE) COMPLEX) | | Descriptor: | 15-cis-1,2-dihydroneurosporene, 2-CHLORO-4-ETHYLAMINO-6-(S(-)-2'-CYANO-4-BUTYLAMINO)-1,3,5-TRIAZINE, BACTERIOCHLOROPHYLL B, ... | | Authors: | Lancaster, C.R.D, Michel, H. | | Deposit date: | 1997-07-31 | | Release date: | 1999-04-06 | | Last modified: | 2023-08-09 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Refined crystal structures of reaction centres from Rhodopseudomonas viridis in complexes with the herbicide atrazine and two chiral atrazine derivatives also lead to a new model of the bound carotenoid.
J.Mol.Biol., 286, 1999
|
|
5PRC
 
 | | PHOTOSYNTHETIC REACTION CENTER FROM RHODOPSEUDOMONAS VIRIDIS (ATRAZINE COMPLEX) | | Descriptor: | 15-cis-1,2-dihydroneurosporene, 2-CHLORO-4-ISOPROPYLAMINO-6-ETHYLAMINO -1,3,5-TRIAZINE, BACTERIOCHLOROPHYLL B, ... | | Authors: | Lancaster, C.R.D, Michel, H. | | Deposit date: | 1997-08-01 | | Release date: | 1999-04-06 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (2.35 Å) | | Cite: | Refined crystal structures of reaction centres from Rhodopseudomonas viridis in complexes with the herbicide atrazine and two chiral atrazine derivatives also lead to a new model of the bound carotenoid.
J.Mol.Biol., 286, 1999
|
|
3PRC
 
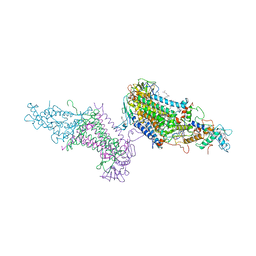 | | PHOTOSYNTHETIC REACTION CENTER FROM RHODOPSEUDOMONAS VIRIDIS (QB-DEPLETED) | | Descriptor: | 15-cis-1,2-dihydroneurosporene, BACTERIOCHLOROPHYLL B, BACTERIOPHEOPHYTIN B, ... | | Authors: | Lancaster, C.R.D, Michel, H. | | Deposit date: | 1997-07-29 | | Release date: | 1998-11-11 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | The coupling of light-induced electron transfer and proton uptake as derived from crystal structures of reaction centres from Rhodopseudomonas viridis modified at the binding site of the secondary quinone, QB.
Structure, 5, 1997
|
|
2IG2
 
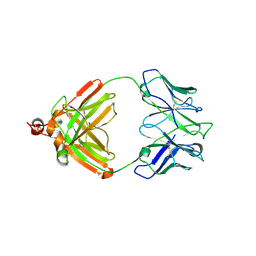 | | DIR PRIMAERSTRUKTUR DES KRISTALLISIERBAREN MONOKLONALEN IMMUNOGLOBULINS IGG1 KOL. II. AMINOSAEURESEQUENZ DER L-KETTE, LAMBDA-TYP, SUBGRUPPE I (GERMAN) | | Descriptor: | IGG1-LAMBDA KOL FAB (HEAVY CHAIN), IGG1-LAMBDA KOL FAB (LIGHT CHAIN) | | Authors: | Marquart, M, Huber, R. | | Deposit date: | 1989-04-18 | | Release date: | 1989-07-12 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | The primary structure of crystallizable monoclonal immunoglobulin IgG1 Kol. II. Amino acid sequence of the L-chain, gamma-type, subgroup I
Biol.Chem.Hoppe-Seyler, 370, 1989
|
|
2JBL
 
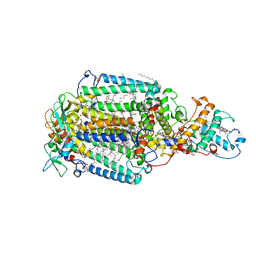 | | PHOTOSYNTHETIC REACTION CENTER FROM BLASTOCHLORIS VIRIDIS | | Descriptor: | 15-cis-1,2-dihydroneurosporene, BACTERIOCHLOROPHYLL B, BACTERIOPHEOPHYTIN B, ... | | Authors: | Lancaster, C.R.D. | | Deposit date: | 2006-12-08 | | Release date: | 2007-03-13 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | A Comparison of Stigmatellin Conformations, Free and Bound to the Photosynthetic Reaction Center and the Cytochrome Bc(1) Complex.
J.Mol.Biol., 368, 2007
|
|
2FB4
 
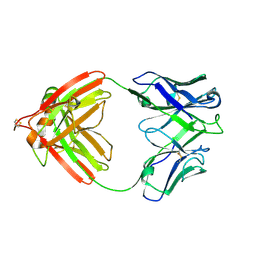 | | DIR PRIMAERSTRUKTUR DES KRISTALLISIERBAREN MONOKLONALEN IMMUNOGLOBULINS IGG1 KOL. II. AMINOSAEURESEQUENZ DER L-KETTE, LAMBDA-TYP, SUBGRUPPE I (GERMAN) | | Descriptor: | IGG1-LAMBDA KOL FAB (HEAVY CHAIN), IGG1-LAMBDA KOL FAB (LIGHT CHAIN) | | Authors: | Marquart, M, Huber, R. | | Deposit date: | 1989-04-18 | | Release date: | 1989-07-12 | | Last modified: | 2024-06-05 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | The primary structure of crystallizable monoclonal immunoglobulin IgG1 Kol. II. Amino acid sequence of the L-chain, gamma-type, subgroup I
Biol.Chem.Hoppe-Seyler, 370, 1989
|
|
3BPS
 
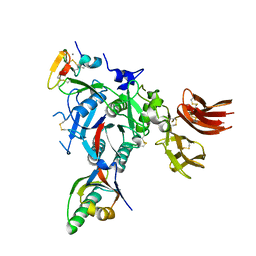 | | PCSK9:EGF-A complex | | Descriptor: | CALCIUM ION, Low-density lipoprotein receptor, Proprotein convertase subtilisin/kexin type 9 | | Authors: | Kwon, H.J. | | Deposit date: | 2007-12-19 | | Release date: | 2008-02-12 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (2.41 Å) | | Cite: | Molecular basis for LDL receptor recognition by PCSK9.
Proc.Natl.Acad.Sci.Usa, 105, 2008
|
|
2PN5
 
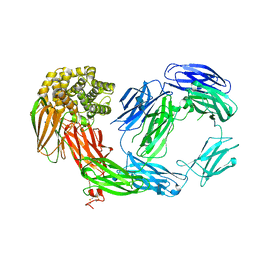 | | Crystal Structure of TEP1r | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, SODIUM ION, ... | | Authors: | Baxter, R.H.G. | | Deposit date: | 2007-04-23 | | Release date: | 2007-07-24 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (2.698 Å) | | Cite: | Structural basis for conserved complement factor-like function in the antimalarial protein TEP1
Proc.Natl.Acad.Sci.Usa, 104, 2007
|
|
1DXR
 
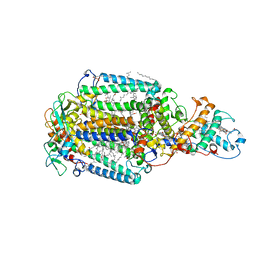 | | Photosynthetic reaction center from Rhodopseudomonas viridis - His L168 Phe mutant (terbutryn complex) | | Descriptor: | 15-cis-1,2-dihydroneurosporene, 2-T-BUTYLAMINO-4-ETHYLAMINO-6-METHYLTHIO-S-TRIAZINE, BACTERIOCHLOROPHYLL B, ... | | Authors: | Lancaster, C.R.D, Bibikova, M, Sabatino, P, Oesterhelt, D, Michel, H. | | Deposit date: | 2000-01-15 | | Release date: | 2001-01-12 | | Last modified: | 2023-12-06 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Structural basis of the drastically increased initial electron transfer rate in the reaction center from a Rhodopseudomonas viridis mutant described at 2.00-A resolution.
J. Biol. Chem., 275, 2000
|
|
8PTI
 
 | | CRYSTAL STRUCTURE OF A Y35G MUTANT OF BOVINE PANCREATIC TRYPSIN INHIBITOR | | Descriptor: | BOVINE PANCREATIC TRYPSIN INHIBITOR | | Authors: | Housset, D, Kim, K.-S, Fuchs, J, Woodward, C, Wlodawer, A. | | Deposit date: | 1990-12-17 | | Release date: | 1991-04-15 | | Last modified: | 2024-06-05 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Crystal structure of a Y35G mutant of bovine pancreatic trypsin inhibitor.
J.Mol.Biol., 220, 1991
|
|
1QLQ
 
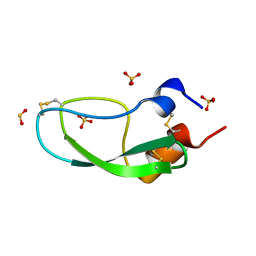 | | Bovine Pancreatic Trypsin Inhibitor (BPTI) Mutant with Altered Binding Loop Sequence | | Descriptor: | PANCREATIC TRYPSIN INHIBITOR, SULFATE ION | | Authors: | Czapinska, H, Krzywda, S, Sheldrick, G.M, Otlewski, J, Jaskolski, M. | | Deposit date: | 1999-09-10 | | Release date: | 1999-10-05 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (1.42 Å) | | Cite: | High Resolution Structure of Bovine Pancreatic Trypsin Inhibitor with Altered Binding Loop Sequence
J.Mol.Biol., 295, 1999
|
|
4RCR
 
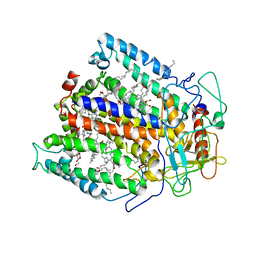 | | STRUCTURE OF THE REACTION CENTER FROM RHODOBACTER SPHAEROIDES R-26 AND 2.4.1: PROTEIN-COFACTOR (BACTERIOCHLOROPHYLL, BACTERIOPHEOPHYTIN, AND CAROTENOID) INTERACTIONS | | Descriptor: | BACTERIOCHLOROPHYLL A, BACTERIOPHEOPHYTIN A, FE (III) ION, ... | | Authors: | Komiya, H, Yeates, T.O, Chirino, A.J, Rees, D.C, Allen, J.P, Feher, G. | | Deposit date: | 1991-09-09 | | Release date: | 1993-10-31 | | Last modified: | 2020-07-29 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Structure of the reaction center from Rhodobacter sphaeroides R-26 and 2.4.1: protein-cofactor (bacteriochlorophyll, bacteriopheophytin, and carotenoid) interactions.
Proc.Natl.Acad.Sci.USA, 85, 1988
|
|
2PTN
 
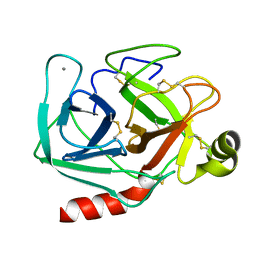 | | ON THE DISORDERED ACTIVATION DOMAIN IN TRYPSINOGEN. CHEMICAL LABELLING AND LOW-TEMPERATURE CRYSTALLOGRAPHY | | Descriptor: | CALCIUM ION, TRYPSIN | | Authors: | Walter, J, Steigemann, W, Singh, T.P, Bartunik, H, Bode, W, Huber, R. | | Deposit date: | 1981-10-26 | | Release date: | 1982-03-04 | | Last modified: | 2024-10-09 | | Method: | X-RAY DIFFRACTION (1.55 Å) | | Cite: | On the Disordered Activation Domain in Trypsinogen. Chemical Labelling and Low-Temperature Crystallography
Acta Crystallogr.,Sect.B, 38, 1982
|
|
4CTS
 
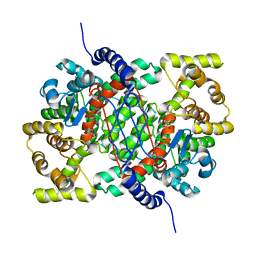 | |
6PTI
 
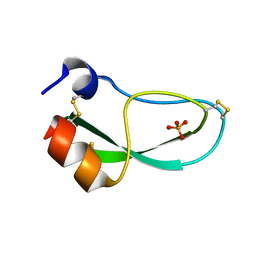 | |
5V8K
 
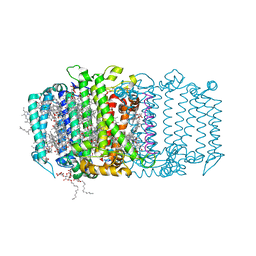 | | Homodimeric reaction center of H. modesticaldum | | Descriptor: | 1-[GLYCEROLYLPHOSPHONYL]-2-[8-(2-HEXYL-CYCLOPROPYL)-OCTANAL-1-YL]-3-[HEXADECANAL-1-YL]-GLYCEROL, 4,4'-Diaponeurosporene, 8(1)-OH-Chlorophyll aF, ... | | Authors: | Gisriel, C, Sarrou, I, Ferlez, B, Golbeck, J, Redding, K.E, Fromme, R. | | Deposit date: | 2017-03-22 | | Release date: | 2017-09-06 | | Last modified: | 2024-03-06 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Structure of a symmetric photosynthetic reaction center-photosystem.
Science, 357, 2017
|
|
2BMH
 
 | |
3BIF
 
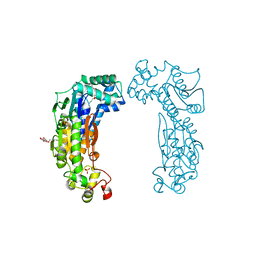 | | 6-PHOSPHOFRUCTO-2-KINASE/FRUCTOSE-2,6-BISPHOSPHATASE EMPTY 6-PF-2K ACTIVE SITE | | Descriptor: | PHOSPHATE ION, PROTEIN (6-PHOSPHOFRUCTO-2-KINASE/ FRUCTOSE-2,6-BISPHOSPHATASE), SUCCINIC ACID, ... | | Authors: | Yuen, M.H, Hasemann, C.A. | | Deposit date: | 1999-05-05 | | Release date: | 1999-09-29 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | A switch in the kinase domain of rat testis 6-phosphofructo-2-kinase/fructose-2,6-bisphosphatase.
Biochemistry, 38, 1999
|
|
1DCE
 
 | |
3GKJ
 
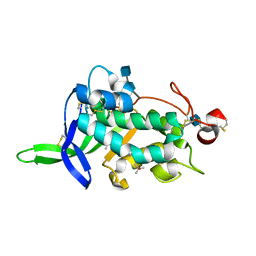 | | NPC1D(NTD):25hydroxycholesterol | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, 25-HYDROXYCHOLESTEROL, ... | | Authors: | Kwon, H.J. | | Deposit date: | 2009-03-10 | | Release date: | 2009-07-14 | | Last modified: | 2021-10-20 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Structure of N-terminal domain of NPC1 reveals distinct subdomains for binding and transfer of cholesterol.
Cell(Cambridge,Mass.), 137, 2009
|
|
3GKH
 
 | | NPC1(NTD) | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, GLYCEROL, ... | | Authors: | Kwon, H.J. | | Deposit date: | 2009-03-10 | | Release date: | 2009-07-14 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (1.81 Å) | | Cite: | Structure of N-terminal domain of NPC1 reveals distinct subdomains for binding and transfer of cholesterol.
Cell(Cambridge,Mass.), 137, 2009
|
|
5PTI
 
 | |
