6E1N
 
 | | Structure of AtTPC1(DDE) in state 1 | | Descriptor: | 1,2-DIACYL-GLYCEROL-3-SN-PHOSPHATE, CALCIUM ION, PALMITIC ACID, ... | | Authors: | Kintzer, A.F, Green, E.M, Cheng, Y, Stroud, R.M. | | Deposit date: | 2018-07-10 | | Release date: | 2018-09-19 | | Last modified: | 2019-12-18 | | Method: | ELECTRON MICROSCOPY (3.7 Å) | | Cite: | Structural basis for activation of voltage sensor domains in an ion channel TPC1.
Proc. Natl. Acad. Sci. U.S.A., 115, 2018
|
|
4ZOU
 
 | |
6E1P
 
 | | Structure of AtTPC1(DDE) in state 2 | | Descriptor: | CALCIUM ION, PALMITIC ACID, Two pore calcium channel protein 1 | | Authors: | Kintzer, A.F, Green, E.M, Cheng, Y, Stroud, R.M. | | Deposit date: | 2018-07-10 | | Release date: | 2018-09-19 | | Last modified: | 2019-12-18 | | Method: | ELECTRON MICROSCOPY (3.7 Å) | | Cite: | Structural basis for activation of voltage sensor domains in an ion channel TPC1.
Proc. Natl. Acad. Sci. U.S.A., 115, 2018
|
|
6E1K
 
 | | Structure of AtTPC1(DDE) reconstituted in saposin A with cat06 Fab | | Descriptor: | CALCIUM ION, PALMITIC ACID, Two pore calcium channel protein 1, ... | | Authors: | Kintzer, A.F, Green, E.M, Cheng, Y, Stroud, R.M. | | Deposit date: | 2018-07-10 | | Release date: | 2018-09-19 | | Last modified: | 2019-12-18 | | Method: | ELECTRON MICROSCOPY (3.3 Å) | | Cite: | Structural basis for activation of voltage sensor domains in an ion channel TPC1.
Proc. Natl. Acad. Sci. U.S.A., 115, 2018
|
|
5BND
 
 | | Crystal structure of the C-terminal domain of TagH | | Descriptor: | ABC transporter, ATP-binding protein | | Authors: | Chen, S.C, Chen, Y. | | Deposit date: | 2015-05-26 | | Release date: | 2016-07-06 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (2.68 Å) | | Cite: | SH3-Like Motif-Containing C-terminal Domain of Staphylococcal Teichoic Acid Transporter Suggests Possible Function.
Proteins, 2016
|
|
7WQA
 
 | | SARS-CoV-2 main protease in complex with Z-VAD-FMK | | Descriptor: | 3C-like proteinase, Z-VAD(OMe)-FMK | | Authors: | Wang, Y.C, Yang, C.S, Hou, M.H, Tsai, C.L, Chen, Y. | | Deposit date: | 2022-01-25 | | Release date: | 2023-03-22 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | SARS-CoV-2 main protease in complex with Z-VAD-FMK
To Be Published
|
|
7WQ9
 
 | | Crystal structure of SARS-CoV-2 main protease in complex with Z-IETD-FMK | | Descriptor: | 3C-like proteinase, Z-IETD-FMK | | Authors: | Wang, Y.C, Yang, C.S, Hou, M.H, Tsai, C.L, Chen, Y. | | Deposit date: | 2022-01-25 | | Release date: | 2023-08-02 | | Method: | X-RAY DIFFRACTION (2.054 Å) | | Cite: | Crystal structure of SARS-CoV-2 main protease in complex with Z-IETD-FMK
To Be Published
|
|
7WQ8
 
 | | Crystal structure of SARS-CoV-2 main protease in complex with Z-DEVD-FMK | | Descriptor: | 3C-like proteinase, Z-DEVD-FMK | | Authors: | Wang, Y.C, Yang, C.S, Hou, M.H, Tsai, C.L, Chen, Y. | | Deposit date: | 2022-01-24 | | Release date: | 2023-08-02 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Crystal structure of SARS-CoV-2 main protease in complex with Z-DEVD-FMK
To Be Published
|
|
7WQB
 
 | | SARS-CoV-2 main protease mutant (P168A) in complex with MG-132 | | Descriptor: | 3C-like proteinase nsp5, MAGNESIUM ION, N-[(benzyloxy)carbonyl]-L-leucyl-N-[(2S)-1-hydroxy-4-methylpentan-2-yl]-L-leucinamide | | Authors: | Wang, Y.C, Yang, C.S, Hou, M.H, Tsai, C.L, Chen, Y. | | Deposit date: | 2022-01-25 | | Release date: | 2023-08-02 | | Last modified: | 2024-11-13 | | Method: | X-RAY DIFFRACTION (1.87 Å) | | Cite: | SARS-CoV-2 main protease mutant (P168A) in complex with MG-132
To Be Published
|
|
7WQK
 
 | | wild-type SARS-CoV-2 main protease in complex with MG-132 | | Descriptor: | 3C-like proteinase, MAGNESIUM ION, N-[(benzyloxy)carbonyl]-L-leucyl-N-[(2S)-1-hydroxy-4-methylpentan-2-yl]-L-leucinamide | | Authors: | Wang, Y.C, Yang, C.S, Hou, M.H, Tsai, C.L, Chen, Y. | | Deposit date: | 2022-01-25 | | Release date: | 2023-08-02 | | Last modified: | 2024-11-13 | | Method: | X-RAY DIFFRACTION (2.15 Å) | | Cite: | wild-type SARS-CoV-2 main protease in complex with MG-132
To Be Published
|
|
4DKE
 
 | | Crystal Structure of Human Interleukin-34 Bound to FAb1.1 | | Descriptor: | FAb1.1 Heavy Chain, FAb1.1 Light Chain, Interleukin-34, ... | | Authors: | Ma, X, Chen, Y, Stawicki, S, Wu, Y, Bazan, J.F, Starovasnik, M.A. | | Deposit date: | 2012-02-03 | | Release date: | 2012-04-11 | | Last modified: | 2020-07-29 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | Structural Basis for the Dual Recognition of Helical Cytokines IL-34 and CSF-1 by CSF-1R.
Structure, 20, 2012
|
|
4DKF
 
 | | Crystal Structure of Human Interleukin-34 Bound to FAb2 | | Descriptor: | FAb2 Heavy Chain, FAb2 Light Chain, Interleukin-34, ... | | Authors: | Ma, X, Chen, Y, Stawicki, S, Wu, Y, Bazan, J.F, Starovasnik, M.A. | | Deposit date: | 2012-02-03 | | Release date: | 2012-04-11 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (2.61 Å) | | Cite: | Structural Basis for the Dual Recognition of Helical Cytokines IL-34 and CSF-1 by CSF-1R.
Structure, 20, 2012
|
|
5A9W
 
 | | Structure of GDPCP BipA | | Descriptor: | GTP-BINDING PROTEIN, PHOSPHOMETHYLPHOSPHONIC ACID GUANYLATE ESTER | | Authors: | Kumar, V, Chen, Y, Ero, R, Li, Z, Gao, Y. | | Deposit date: | 2015-07-23 | | Release date: | 2015-08-26 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (3.7 Å) | | Cite: | Structure of Bipa in GTP Form Bound to the Ratcheted Ribosome.
Proc.Natl.Acad.Sci.USA, 112, 2015
|
|
7YBF
 
 | |
4E0R
 
 | | Structure of the chicken MHC class I molecule BF2*0401 | | Descriptor: | 8-MERIC PEPTIDE (FUS/TLS), Beta-2 microglobulin, MHC class I alpha chain 2 | | Authors: | Zhang, J, Chen, Y, Qi, J, Gao, F, Kaufman, J, Xia, C, Gao, G.F. | | Deposit date: | 2012-03-05 | | Release date: | 2012-11-21 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (2.26 Å) | | Cite: | Narrow Groove and Restricted Anchors of MHC Class I Molecule BF2*0401 Plus Peptide Transporter Restriction Can Explain Disease Susceptibility of B4 Chickens.
J.Immunol., 189, 2012
|
|
2KGP
 
 | | Structural basis for stabilization of the tau pre-mRNA splicing regulatory element by Novantrone (Mitoxantrone) | | Descriptor: | 1,4-DIHYDROXY-5,8-BIS({2-[(2-HYDROXYETHYL)AMINO]ETHYL}AMINO)-9,10-ANTHRACENEDIONE, RNA (25-MER) | | Authors: | Zheng, S, Chen, Y, Donahue, C.P, Wolfe, M.S, Varani, G. | | Deposit date: | 2009-03-13 | | Release date: | 2009-06-02 | | Last modified: | 2024-05-01 | | Method: | SOLUTION NMR | | Cite: | Structural basis for stabilization of the tau pre-mRNA splicing regulatory element by novantrone (mitoxantrone).
Chem.Biol., 16, 2009
|
|
5A9V
 
 | | Structure of apo BipA | | Descriptor: | GTP-BINDING PROTEIN | | Authors: | Kumar, V, Chen, Y, Ero, R, Li, Z, Gao, Y. | | Deposit date: | 2015-07-23 | | Release date: | 2015-09-02 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (3.31 Å) | | Cite: | Structure of Bipa in GTP Form Bound to the Ratcheted Ribosome.
Proc.Natl.Acad.Sci.USA, 112, 2015
|
|
4G43
 
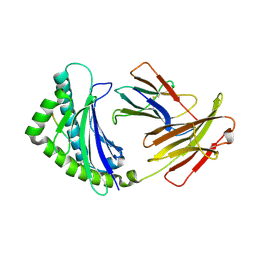 | | Structure of the chicken MHC class I molecule BF2*0401 complexed to P5E | | Descriptor: | (4S)-2-METHYL-2,4-PENTANEDIOL, 8-MERIC PEPTIDE P5E, Beta-2 microglobulin, ... | | Authors: | Zhang, J, Chen, Y, Qi, J, Gao, F, Liu, J, Kaufman, J, Xia, C, Gao, G.F. | | Deposit date: | 2012-07-16 | | Release date: | 2012-11-21 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (1.803 Å) | | Cite: | Narrow Groove and Restricted Anchors of MHC Class I Molecule BF2*0401 Plus Peptide Transporter Restriction Can Explain Disease Susceptibility of B4 Chickens.
J.Immunol., 189, 2012
|
|
1XI4
 
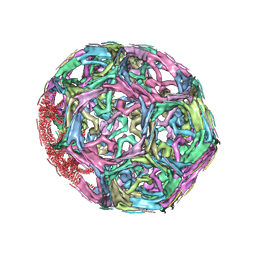 | | Clathrin D6 Coat | | Descriptor: | Clathrin heavy chain, Clathrin light chain A | | Authors: | Fotin, A, Cheng, Y, Sliz, P, Grigorieff, N, Harrison, S.C, Kirchhausen, T, Walz, T. | | Deposit date: | 2004-09-21 | | Release date: | 2004-11-02 | | Last modified: | 2024-03-13 | | Method: | ELECTRON MICROSCOPY (7.9 Å) | | Cite: | Molecular model for a complete clathrin lattice from electron cryomicroscopy
Nature, 432, 2004
|
|
2HZB
 
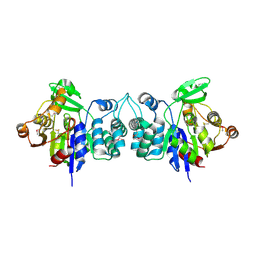 | | X-Ray Crystal Structure of Protein BH3568 from Bacillus halodurans. Northeast Structural Genomics Consortium BhR60. | | Descriptor: | Hypothetical UPF0052 protein BH3568 | | Authors: | Kuzin, A.P, Chen, Y, Seetharaman, J, Benach, J, Shastry, R, Conover, K, Ma, L.C, Xiao, R, Liu, J, Baran, M.C, Acton, T.B, Rost, B, Montelione, G.T, Hunt, J.F, Tong, L, Northeast Structural Genomics Consortium (NESG) | | Deposit date: | 2006-08-08 | | Release date: | 2006-08-22 | | Last modified: | 2024-11-13 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | X-Ray structure of the hypothetical UPF0052 protein BH3568 from Bacillus halodurans. Northeast Structural Genomics Consortium BhR60.
To be Published
|
|
5YRP
 
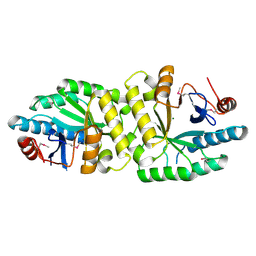 | | Crystal structure of the EAL domain of Mycobacterium smegmatis DcpA | | Descriptor: | MAGNESIUM ION, Sensory box/response regulator | | Authors: | Chen, H.J, li, N, Luo, Y, Jiang, Y.L, Zhou, C.Z, Chen, Y, Li, Q. | | Deposit date: | 2017-11-09 | | Release date: | 2018-05-09 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (2.99 Å) | | Cite: | The GDP-switched GAF domain of DcpA modulates the concerted synthesis/hydrolysis of c-di-GMP inMycobacterium smegmatis.
Biochem. J., 475, 2018
|
|
5XP7
 
 | | C-Src in complex with ATP-CHCl | | Descriptor: | GLYCEROL, MAGNESIUM ION, Proto-oncogene tyrosine-protein kinase Src, ... | | Authors: | Guo, M, Dai, S, Duan, Y, Chen, L, Chen, Y. | | Deposit date: | 2017-06-01 | | Release date: | 2017-09-06 | | Last modified: | 2024-03-27 | | Method: | X-RAY DIFFRACTION (2.012 Å) | | Cite: | Remarkably Stereospecific Utilization of ATP alpha , beta-Halomethylene Analogues by Protein Kinases.
J. Am. Chem. Soc., 139, 2017
|
|
7YIB
 
 | | Crystal structure of wild-type Cap4 SAVED domain-containing receptor from Enterobacter cloacae | | Descriptor: | CD-NTase-associated protein 4, CHLORIDE ION | | Authors: | Ko, T.-P, Yang, C.-S, Hou, M.-H, Wang, Y.-C, Chen, Y. | | Deposit date: | 2022-07-15 | | Release date: | 2023-05-31 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (2.18 Å) | | Cite: | Specific recognition of cyclic oligonucleotides by Cap4 for phage infection.
Int.J.Biol.Macromol., 237, 2023
|
|
6B0Y
 
 | | Crystal Structure of small molecule ARS-917 covalently bound to K-Ras G12C | | Descriptor: | 1-{4-[6-chloro-7-(2-fluorophenyl)quinazolin-4-yl]piperazin-1-yl}propan-1-one, CALCIUM ION, GLYCEROL, ... | | Authors: | Hansen, R, Peters, U, Babbar, A, Chen, Y, Feng, J, Janes, M.R, Li, L.-S, Ren, P, Liu, Y, Zarrinkar, P.P. | | Deposit date: | 2017-09-15 | | Release date: | 2018-05-16 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (1.43 Å) | | Cite: | The reactivity-driven biochemical mechanism of covalent KRASG12Cinhibitors.
Nat. Struct. Mol. Biol., 25, 2018
|
|
7YIA
 
 | | Crystal structure of K74A mutant of Cap4 SAVED domain-containing receptor from Enterobacter cloacae | | Descriptor: | CD-NTase-associated protein 4, MAGNESIUM ION | | Authors: | Ko, T.-P, Yang, C.-S, Hou, M.-H, Wang, Y.-C, Chen, Y. | | Deposit date: | 2022-07-15 | | Release date: | 2023-05-31 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.428 Å) | | Cite: | Specific recognition of cyclic oligonucleotides by Cap4 for phage infection.
Int.J.Biol.Macromol., 237, 2023
|
|
