2G3J
 
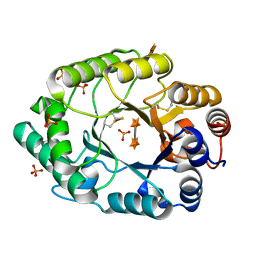 | | Structure of S.olivaceoviridis xylanase Q88A/R275A mutant | | Descriptor: | PHOSPHATE ION, Xylanase, alpha-D-xylopyranose-(1-4)-alpha-D-xylopyranose | | Authors: | Diertavitian, S, Kaneko, S, Fujimoto, Z, Kuno, A, Johansson, E, Lo Leggio, L. | | Deposit date: | 2006-02-20 | | Release date: | 2007-03-06 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Structure-based engineering of glucose specificity in a family 10 xylanase from Streptomyces olivaceoviridis E-86
PROCESS BIOCHEM, 47, 2012
|
|
5I20
 
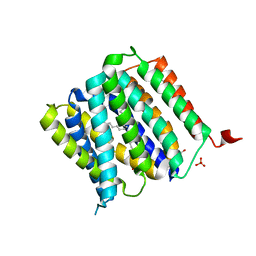 | | Crystal structure of protein | | Descriptor: | (2R)-2,3-dihydroxypropyl (9Z)-octadec-9-enoate, SULFATE ION, Uncharacterized protein | | Authors: | Ishitani, R, Nureki, O. | | Deposit date: | 2016-02-08 | | Release date: | 2016-06-01 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Structural basis for amino acid export by DMT superfamily transporter YddG.
Nature, 534, 2016
|
|
7VED
 
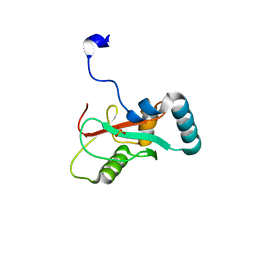 | |
7VEC
 
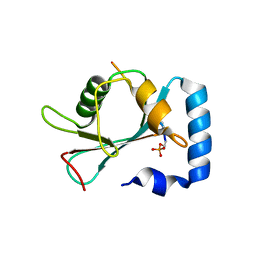 | |
6AAJ
 
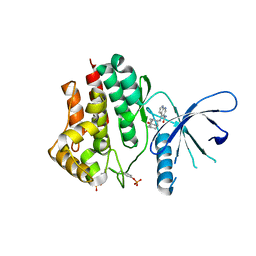 | | Crystal structure of JAK2 in complex with peficitinib | | Descriptor: | 4-[[(1S,3R)-5-oxidanyl-2-adamantyl]amino]-1H-pyrrolo[2,3-b]pyridine-5-carboxamide, Tyrosine-protein kinase JAK2 | | Authors: | Amano, Y, Tateishi, Y. | | Deposit date: | 2018-07-18 | | Release date: | 2018-08-15 | | Last modified: | 2023-03-08 | | Method: | X-RAY DIFFRACTION (2.37 Å) | | Cite: | Discovery and structural characterization of peficitinib (ASP015K) as a novel and potent JAK inhibitor
Bioorg. Med. Chem., 26, 2018
|
|
6AAM
 
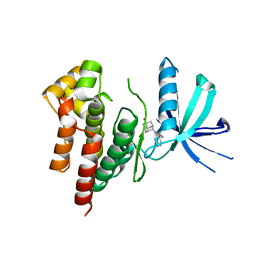 | | Crystal structure of TYK2 in complex with peficitinib | | Descriptor: | 4-[[(1S,3R)-5-oxidanyl-2-adamantyl]amino]-1H-pyrrolo[2,3-b]pyridine-5-carboxamide, Non-receptor tyrosine-protein kinase TYK2 | | Authors: | Nomura, N, Tomimoto, Y. | | Deposit date: | 2018-07-18 | | Release date: | 2018-08-15 | | Last modified: | 2024-03-27 | | Method: | X-RAY DIFFRACTION (1.98 Å) | | Cite: | Discovery and structural characterization of peficitinib (ASP015K) as a novel and potent JAK inhibitor
Bioorg. Med. Chem., 26, 2018
|
|
6AAH
 
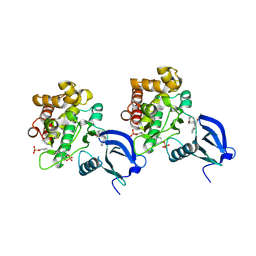 | | Crystal structure of JAK1 in complex with peficitinib | | Descriptor: | 4-[[(1S,3R)-5-oxidanyl-2-adamantyl]amino]-1H-pyrrolo[2,3-b]pyridine-5-carboxamide, Tyrosine-protein kinase JAK1 | | Authors: | Amano, Y. | | Deposit date: | 2018-07-18 | | Release date: | 2018-08-15 | | Last modified: | 2018-10-24 | | Method: | X-RAY DIFFRACTION (1.83 Å) | | Cite: | Discovery and structural characterization of peficitinib (ASP015K) as a novel and potent JAK inhibitor
Bioorg. Med. Chem., 26, 2018
|
|
6AAK
 
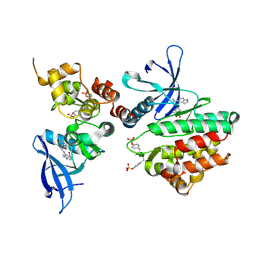 | | Crystal structure of JAK3 in complex with peficitinib | | Descriptor: | 4-[[(1S,3R)-5-oxidanyl-2-adamantyl]amino]-1H-pyrrolo[2,3-b]pyridine-5-carboxamide, Tyrosine-protein kinase JAK3 | | Authors: | Amano, Y. | | Deposit date: | 2018-07-18 | | Release date: | 2018-08-15 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (2.67 Å) | | Cite: | Discovery and structural characterization of peficitinib (ASP015K) as a novel and potent JAK inhibitor
Bioorg. Med. Chem., 26, 2018
|
|
1HMK
 
 | | RECOMBINANT GOAT ALPHA-LACTALBUMIN | | Descriptor: | CALCIUM ION, PROTEIN (ALPHA-LACTALBUMIN) | | Authors: | Horii, K, Matsushima, M, Tsumoto, K, Kumagai, I. | | Deposit date: | 1998-11-26 | | Release date: | 1999-11-26 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Effect of the extra n-terminal methionine residue on the stability and folding of recombinant alpha-lactalbumin expressed in Escherichia coli.
J.Mol.Biol., 285, 1999
|
|
1H2A
 
 | | SINGLE CRYSTALS OF HYDROGENASE FROM DESULFOVIBRIO VULGARIS | | Descriptor: | FE3-S4 CLUSTER, HYDROGENASE, IRON/SULFUR CLUSTER, ... | | Authors: | Higuchi, Y, Yasuoka, N. | | Deposit date: | 1997-10-17 | | Release date: | 1999-02-09 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Unusual ligand structure in Ni-Fe active center and an additional Mg site in hydrogenase revealed by high resolution X-ray structure analysis.
Structure, 5, 1997
|
|
7QDA
 
 | | Crystal structure of CalpL | | Descriptor: | CalpL, SULFATE ION, TETRAETHYLENE GLYCOL, ... | | Authors: | Schneberger, N, Hagelueken, G. | | Deposit date: | 2021-11-26 | | Release date: | 2022-11-16 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Antiviral signalling by a cyclic nucleotide activated CRISPR protease.
Nature, 614, 2023
|
|
7V2Z
 
 | | ZIKV NS3helicase in complex with ssRNA and ATP-Mn2+ | | Descriptor: | ADENOSINE-5'-TRIPHOSPHATE, Core protein, MANGANESE (II) ION, ... | | Authors: | Lin, M.M, Yang, H.T. | | Deposit date: | 2021-08-10 | | Release date: | 2022-08-17 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.10101676 Å) | | Cite: | Structural Basis of Zika Virus Helicase in RNA Unwinding and ATP Hydrolysis.
Acs Infect Dis., 8, 2022
|
|
8B0R
 
 | | Structure of the CalpL/cA4 complex | | Descriptor: | Cyclic tetraadenosine monophosphate (cA4), SMODS-associated and fused to various effectors domain-containing protein, SULFATE ION, ... | | Authors: | Schneberger, N, Hagelueken, G. | | Deposit date: | 2022-09-08 | | Release date: | 2022-11-16 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Antiviral signalling by a cyclic nucleotide activated CRISPR protease.
Nature, 614, 2023
|
|
8B0U
 
 | | Structure of the CalpL/T10 complex | | Descriptor: | CalpT10, GLYCEROL, SAVED domain-containing protein, ... | | Authors: | Schneberger, N, Hagelueken, G. | | Deposit date: | 2022-09-08 | | Release date: | 2022-11-16 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (3.29 Å) | | Cite: | Antiviral signalling by a cyclic nucleotide activated CRISPR protease.
Nature, 614, 2023
|
|
7WEB
 
 | |
7WE9
 
 | | SARS-CoV-2 Omicron variant spike protein in complex with Fab XGv289 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Spike glycoprotein, ... | | Authors: | Wang, X, Wang, L. | | Deposit date: | 2021-12-23 | | Release date: | 2022-05-04 | | Method: | ELECTRON MICROSCOPY (3.6 Å) | | Cite: | Memory B cell repertoire from triple vaccinees against diverse SARS-CoV-2 variants.
Nature, 603, 2022
|
|
7WLC
 
 | |
7WED
 
 | |
7WEA
 
 | |
7WEF
 
 | |
7WE8
 
 | | SARS-CoV-2 Omicron variant spike protein in complex with Fab XGv265 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Heavy chain of Fab 265, ... | | Authors: | Wang, X, Wang, L. | | Deposit date: | 2021-12-23 | | Release date: | 2022-05-04 | | Method: | ELECTRON MICROSCOPY (3.5 Å) | | Cite: | Memory B cell repertoire from triple vaccinees against diverse SARS-CoV-2 variants.
Nature, 603, 2022
|
|
7WEC
 
 | |
7WE7
 
 | | SARS-CoV-2 Omicron variant spike protein in complex with Fab XGv282 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Heavy chain of Fab 282, ... | | Authors: | Wang, X, Wang, L. | | Deposit date: | 2021-12-23 | | Release date: | 2022-05-04 | | Last modified: | 2024-10-16 | | Method: | ELECTRON MICROSCOPY (3.8 Å) | | Cite: | Memory B cell repertoire from triple vaccinees against diverse SARS-CoV-2 variants.
Nature, 603, 2022
|
|
7WEE
 
 | |
7C8Q
 
 | | Blasnase-T13A with D-asn | | Descriptor: | Asparaginase, D-ASPARAGINE, FORMIC ACID, ... | | Authors: | Lu, F, Ran, T, Jiao, L, Wang, W. | | Deposit date: | 2020-06-03 | | Release date: | 2021-06-09 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (1.89 Å) | | Cite: | Structures of l-asparaginase from Bacillus licheniformis Reveal an Essential Residue for its Substrate Stereoselectivity.
J.Agric.Food Chem., 69, 2021
|
|
