7VTF
 
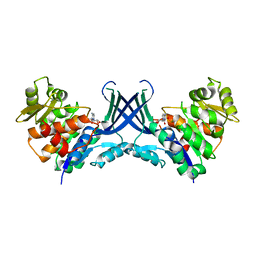 | |
7VTD
 
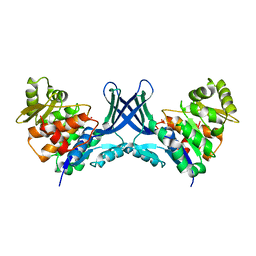 | |
7VTE
 
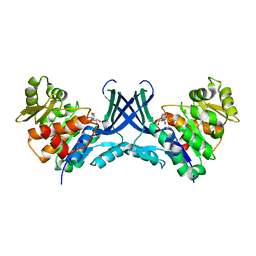 | |
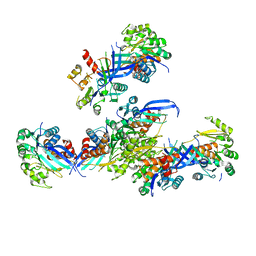
7VVA
 
 | | Pseudouridine bound structure of Pseudouridine kinase (PUKI) from Escherichia coli strain B | | Descriptor: | 5-[(2~{S},3~{R},4~{S},5~{R})-5-(hydroxymethyl)-3,4-bis(oxidanyl)oxolan-2-yl]-1~{H}-pyrimidine-2,4-dione, Pseudouridine kinase | | Authors: | Kim, S.H, Rhee, S. | | Deposit date: | 2021-11-05 | | Release date: | 2022-04-06 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.75029182 Å) | | Cite: | Substrate-binding loop interactions with pseudouridine trigger conformational changes that promote catalytic efficiency of pseudouridine kinase PUKI.
J.Biol.Chem., 298, 2022
|
|
7VTG
 
 | | Pseudouridine bound structure of Pseudouridine kinase (PUKI) S30A mutant from Escherichia coli strain B | | Descriptor: | 5-[(2~{S},3~{R},4~{S},5~{R})-5-(hydroxymethyl)-3,4-bis(oxidanyl)oxolan-2-yl]-1~{H}-pyrimidine-2,4-dione, Pseudouridine kinase | | Authors: | Kim, S.H, Rhee, S. | | Deposit date: | 2021-10-29 | | Release date: | 2022-04-06 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (1.89859128 Å) | | Cite: | Substrate-binding loop interactions with pseudouridine trigger conformational changes that promote catalytic efficiency of pseudouridine kinase PUKI.
J.Biol.Chem., 298, 2022
|
|
3K2D
 
 | |
5XU6
 
 | | Crystal structure of inositol 1,3,4,5,6-pentakisphosphate 2-kinase (IPK1) from Cryptococcus neoformans | | Descriptor: | Inositol-pentakisphosphate 2-kinase, SULFATE ION | | Authors: | Oh, J, Rhee, S. | | Deposit date: | 2017-06-22 | | Release date: | 2017-10-04 | | Last modified: | 2017-11-29 | | Method: | X-RAY DIFFRACTION (2.35 Å) | | Cite: | Crystal structure of inositol 1,3,4,5,6-pentakisphosphate 2-kinase from Cryptococcus neoformans.
J. Struct. Biol., 200, 2017
|
|
3OUM
 
 | | Crystal Structure of toxoflavin-degrading enzyme in complex with toxoflavin | | Descriptor: | 1,6-dimethylpyrimido[5,4-e][1,2,4]triazine-5,7(1H,6H)-dione, MANGANESE (II) ION, toxoflavin-degrading enzyme | | Authors: | Kim, M.I, Rhee, S. | | Deposit date: | 2010-09-15 | | Release date: | 2011-08-10 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Structural and functional analysis of phytotoxin toxoflavin-degrading enzyme
Plos One, 6, 2011
|
|
3OUL
 
 | |
3CZG
 
 | | Crystal Structure Analysis of Sucrose hydrolase (SUH)-glucose complex | | Descriptor: | Sucrose hydrolase, alpha-D-glucopyranose | | Authors: | Kim, M.I, Rhee, S. | | Deposit date: | 2008-04-29 | | Release date: | 2008-07-15 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Crystal structures and mutagenesis of sucrose hydrolase from Xanthomonas axonopodis pv. glycines: insight into the exclusively hydrolytic amylosucrase fold.
J.Mol.Biol., 380, 2008
|
|
3CZE
 
 | | Crystal Structure Analysis of Sucrose hydrolase (SUH)- Tris complex | | Descriptor: | 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, Sucrose hydrolase | | Authors: | Kim, M.I, Rhee, S. | | Deposit date: | 2008-04-29 | | Release date: | 2008-07-15 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Crystal structures and mutagenesis of sucrose hydrolase from Xanthomonas axonopodis pv. glycines: insight into the exclusively hydrolytic amylosucrase fold.
J.Mol.Biol., 380, 2008
|
|
3CZK
 
 | | Crystal Structure Analysis of Sucrose hydrolase(SUH) E322Q-sucrose complex | | Descriptor: | Sucrose hydrolase, beta-D-fructofuranose-(2-1)-alpha-D-glucopyranose | | Authors: | Kim, M.I, Rhee, S. | | Deposit date: | 2008-04-29 | | Release date: | 2008-07-15 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Crystal structures and mutagenesis of sucrose hydrolase from Xanthomonas axonopodis pv. glycines: insight into the exclusively hydrolytic amylosucrase fold.
J.Mol.Biol., 380, 2008
|
|
3CZL
 
 | |
4OK7
 
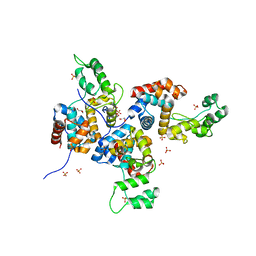 | | Structure of bacteriophage SPN1S endolysin from Salmonella typhimurium | | Descriptor: | Endolysin, GLYCEROL, SULFATE ION | | Authors: | Park, Y, Lim, J, Kong, M, Ryu, S, Rhee, S. | | Deposit date: | 2014-01-22 | | Release date: | 2014-03-19 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Structure of bacteriophage SPN1S endolysin reveals an unusual two-module fold for the peptidoglycan lytic and binding activity.
Mol.Microbiol., 92, 2014
|
|
4P5F
 
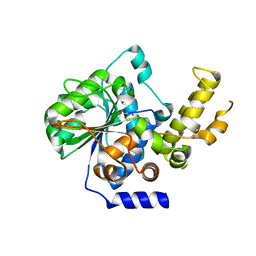 | | The crystal structure of type III effector protein XopQ complexed with adenosine diphosphate ribose | | Descriptor: | CALCIUM ION, Inosine-uridine nucleoside N-ribohydrolase, [(2R,3S,4R,5R)-5-(6-AMINOPURIN-9-YL)-3,4-DIHYDROXY-OXOLAN-2-YL]METHYL [HYDROXY-[[(2R,3S,4R,5S)-3,4,5-TRIHYDROXYOXOLAN-2-YL]METHOXY]PHOSPHORYL] HYDROGEN PHOSPHATE | | Authors: | Yu, S, Hwang, I, Rhee, S. | | Deposit date: | 2014-03-17 | | Release date: | 2014-08-13 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | The crystal structure of type III effector protein XopQ from Xanthomonas oryzae complexed with adenosine diphosphate ribose.
Proteins, 82, 2014
|
|
3DUL
 
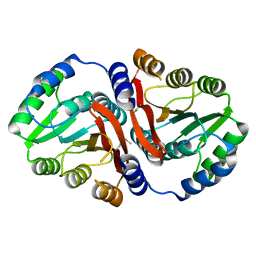 | |
3DUW
 
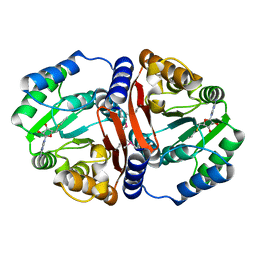 | |
2HK0
 
 | | Crystal structure of D-psicose 3-epimerase (DPEase) in the absence of substrate | | Descriptor: | D-PSICOSE 3-EPIMERASE | | Authors: | Kim, K, Kim, H.J, Oh, D.K, Cha, S.S, Rhee, S. | | Deposit date: | 2006-07-03 | | Release date: | 2006-08-29 | | Last modified: | 2017-10-18 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Crystal Structure of d-Psicose 3-epimerase from Agrobacterium tumefaciens and its Complex with True Substrate d-Fructose: A Pivotal Role of Metal in Catalysis, an Active Site for the Non-phosphorylated Substrate, and its Conformational Changes
J.Mol.Biol., 361, 2006
|
|
3HQ0
 
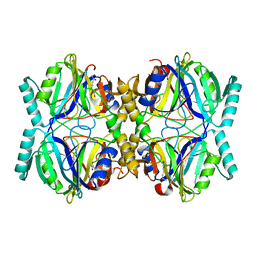 | | Crystal Structure Analysis of the 2,3-dioxygenase LapB from Pseudomonas in complex with a product | | Descriptor: | (2E,4E)-2-hydroxy-6-oxohepta-2,4-dienoic acid, Catechol 2,3-dioxygenase, FE (III) ION | | Authors: | Cho, J.-H, Rhee, S. | | Deposit date: | 2009-06-05 | | Release date: | 2009-10-20 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Crystal structure and functional analysis of the extradiol dioxygenase LapB from a long-chain alkylphenol degradation pathway in Pseudomonas
J.Biol.Chem., 284, 2009
|
|
3HPV
 
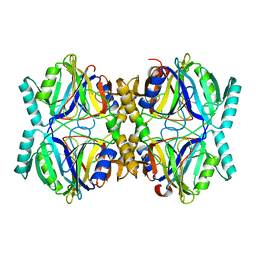 | |
3HPY
 
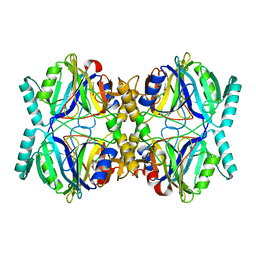 | |
2HK1
 
 | | Crystal structure of D-psicose 3-epimerase (DPEase) in the presence of D-fructose | | Descriptor: | D-PSICOSE 3-EPIMERASE, D-fructose, MANGANESE (II) ION | | Authors: | Kim, K, Kim, H.J, Oh, D.K, Cha, S.S, Rhee, S. | | Deposit date: | 2006-07-03 | | Release date: | 2006-08-29 | | Last modified: | 2017-10-18 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Crystal Structure of d-Psicose 3-epimerase from Agrobacterium tumefaciens and its Complex with True Substrate d-Fructose: A Pivotal Role of Metal in Catalysis, an Active Site for the Non-phosphorylated Substrate, and its Conformational Changes
J.Mol.Biol., 361, 2006
|
|
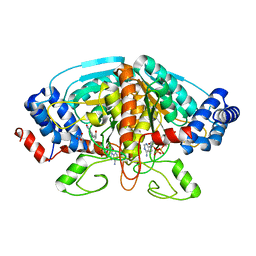
4FJU
 
 | |
4FJS
 
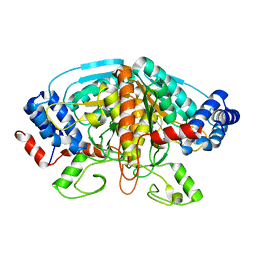 | | Crystal structure of ureidoglycolate dehydrogenase enzyme in apo form | | Descriptor: | Ureidoglycolate dehydrogenase | | Authors: | Kim, M.I, Shin, I, Lee, J, Rhee, S. | | Deposit date: | 2012-06-12 | | Release date: | 2013-01-16 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (2.13 Å) | | Cite: | Structural and functional insights into (s)-ureidoglycolate dehydrogenase, a metabolic branch point enzyme in nitrogen utilization.
Plos One, 7, 2012
|
|
4NNC
 
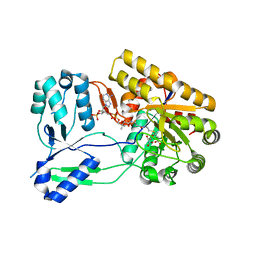 | | Ternary complex of ObcA with C4-CoA adduct and oxalate | | Descriptor: | (3S)-3-[2-[3-[[(2R)-4-[[[(2R,3S,4R,5R)-5-(6-aminopurin-9-yl)-4-oxidanyl-3-phosphonooxy-oxolan-2-yl]methoxy-oxidanyl-phosphoryl]oxy-oxidanyl-phosphoryl]oxy-3,3-dimethyl-2-oxidanyl-butanoyl]amino]propanoylamino]ethylsulfanyl]-3-oxidanyl-butanoic acid, COBALT (II) ION, OBCA, ... | | Authors: | Oh, J.T, Goo, E, Hwang, I, Rhee, S. | | Deposit date: | 2013-11-17 | | Release date: | 2014-03-19 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (2.279 Å) | | Cite: | Structural Basis for Bacterial Quorum Sensing-mediated Oxalogenesis.
J.Biol.Chem., 289, 2014
|
|
