1G76
 
 | | X-RAY STRUCTURE OF ESCHERICHIA COLI PYRIDOXINE 5'-PHOSPHATE OXIDASE COMPLEXED WITH PYRIDOXAL 5'-PHOSPHATE AT 2.0 A RESOLUTION | | Descriptor: | FLAVIN MONONUCLEOTIDE, PHOSPHATE ION, PYRIDOXAL-5'-PHOSPHATE, ... | | Authors: | Safo, M.K, Musayev, F.N, di Salvo, M.L, Schirch, V. | | Deposit date: | 2000-11-09 | | Release date: | 2000-11-29 | | Last modified: | 2023-08-09 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | X-ray structure of Escherichia coli pyridoxine 5'-phosphate oxidase complexed with pyridoxal 5'-phosphate at 2.0 A resolution.
J.Mol.Biol., 310, 2001
|
|
1JNW
 
 | | Active Site Structure of E. coli pyridoxine 5'-phosphate Oxidase | | Descriptor: | FLAVIN MONONUCLEOTIDE, PHOSPHATE ION, PYRIDOXAL-5'-PHOSPHATE, ... | | Authors: | di Salvo, M.L, Ko, T.P, Musayev, F.N, Raboni, S, Schirch, V, Safo, M.K. | | Deposit date: | 2001-07-25 | | Release date: | 2001-08-01 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (2.07 Å) | | Cite: | Active site structure and stereospecificity of Escherichia coli pyridoxine-5'-phosphate oxidase.
J.Mol.Biol., 315, 2002
|
|
1QXD
 
 | | Structural Basis for the Potent Antisickling Effect of a Novel Class of 5-Membered Heterocyclic Aldehydic Compounds | | Descriptor: | FURFURAL, Hemoglobin alpha chain, Hemoglobin beta chain, ... | | Authors: | Safo, M.K, Abdulmalik, O, Danso-Danquah, R, Nokuri, S, Joshi, G.S, Musayev, F.N, Asakura, T, Abraham, D.J. | | Deposit date: | 2003-09-05 | | Release date: | 2003-09-16 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (2.25 Å) | | Cite: | Structural basis for the potent antisickling effect of a novel class of five-membered heterocyclic aldehydic compounds
J.Med.Chem., 47, 2004
|
|
1QXE
 
 | | Structural Basis for the Potent Antisickling Effect of a Novel Class of 5-Membered Heterocyclic Aldehydic Compounds | | Descriptor: | 5-HYDROXYMETHYL-FURFURAL, Hemoglobin alpha chain, Hemoglobin beta chain, ... | | Authors: | Safo, M.K, Abdulmalik, O, Danso-Danquah, R, Nokuri, S, Joshi, G.S, Musayev, F.N, Asakura, T, Abraham, D.J. | | Deposit date: | 2003-09-05 | | Release date: | 2003-09-16 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | Structural basis for the potent antisickling effect of a novel class of five-membered heterocyclic aldehydic compounds
J.Med.Chem., 47, 2004
|
|
1RVU
 
 | | E75Q MUTANT OF RABBIT CYTOSOLIC SERINE HYDROXYMETHYLTRANSFERASE | | Descriptor: | PHOSPHATE ION, PYRIDOXAL-5'-PHOSPHATE, Serine hydroxymethyltransferase, ... | | Authors: | Szebenyi, D.M, Musayev, F.N, Di Salvo, M.L, Safo, M.K, Schirch, V. | | Deposit date: | 2003-12-15 | | Release date: | 2004-06-15 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Serine Hydroxymethyltransferase: Role of Glu75 and Evidence that Serine Is Cleaved by a Retroaldol Mechanism.
Biochemistry, 43, 2004
|
|
1SD4
 
 | | Crystal Structure of a SeMet derivative of BlaI at 2.0 A | | Descriptor: | PENICILLINASE REPRESSOR, SULFATE ION | | Authors: | Safo, M.K, Zhao, Q, Musayev, F.N, Robinson, H, Scarsdale, N, Archer, G.L. | | Deposit date: | 2004-02-13 | | Release date: | 2004-08-10 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Crystal structures of the BlaI repressor from Staphylococcus aureus and its complex with DNA: insights into transcriptional regulation of the bla and mec operons
J.Bacteriol., 187, 2005
|
|
1SD6
 
 | | Crystal Structure of Native MecI at 2.65 A | | Descriptor: | Methicillin resistance regulatory protein mecI | | Authors: | Safo, M.K, Zhao, Q, Musayev, F.N, Robinson, H, Scarsdale, N, Archer, G.L. | | Deposit date: | 2004-02-13 | | Release date: | 2004-02-24 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2.65 Å) | | Cite: | Crystal structures of the BlaI repressor from Staphylococcus aureus and its complex with DNA: insights into transcriptional regulation of the bla and mec operons
J.Bacteriol., 187, 2005
|
|
1RVY
 
 | | E75Q MUTANT OF RABBIT CYTOSOLIC SERINE HYDROXYMETHYLTRANSFERASE, COMPLEX WITH GLYCINE | | Descriptor: | N-GLYCINE-[3-HYDROXY-2-METHYL-5-PHOSPHONOOXYMETHYL-PYRIDIN-4-YL-METHANE], PHOSPHATE ION, PYRIDOXAL-5'-PHOSPHATE, ... | | Authors: | Szebenyi, D.M, Musayev, F.N, Di Salvo, M.L, Safo, M.K, Schirch, V. | | Deposit date: | 2003-12-15 | | Release date: | 2004-06-15 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (2.9 Å) | | Cite: | Serine Hydroxymethyltransferase: Role of Glu75 and Evidence that Serine Is Cleaved by a Retroaldol Mechanism.
Biochemistry, 43, 2004
|
|
1SD7
 
 | | Crystal Structure of a SeMet derivative of MecI at 2.65 A | | Descriptor: | Methicillin resistance regulatory protein mecI | | Authors: | Safo, M.K, Zhao, Q, Musayev, F.N, Robinson, H, Scarsdale, N, Archer, G.L. | | Deposit date: | 2004-02-13 | | Release date: | 2004-02-24 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (2.65 Å) | | Cite: | Crystal structures of the BlaI repressor from Staphylococcus aureus and its complex with DNA: insights into transcriptional regulation of the bla and mec operons
J.Bacteriol., 187, 2005
|
|
1RV4
 
 | | E75L MUTANT OF RABBIT CYTOSOLIC SERINE HYDROXYMETHYLTRANSFERASE | | Descriptor: | PHOSPHATE ION, PYRIDOXAL-5'-PHOSPHATE, Serine hydroxymethyltransferase, ... | | Authors: | Szebenyi, D.M, Musayev, F.N, Di Salvo, M.L, Safo, M.K, Schirch, V. | | Deposit date: | 2003-12-12 | | Release date: | 2004-06-15 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (2.95 Å) | | Cite: | Serine Hydroxymethyltransferase: Role of Glu75 and Evidence that Serine Is Cleaved by a Retroaldol Mechanism.
Biochemistry, 43, 2004
|
|
1QQW
 
 | | CRYSTAL STRUCTURE OF HUMAN ERYTHROCYTE CATALASE | | Descriptor: | CATALASE, PROTOPORPHYRIN IX CONTAINING FE | | Authors: | Ko, T.P, Safo, M.K, Musayev, F.N, Wang, C, Wu, S.H, Abraham, D.J. | | Deposit date: | 1999-06-09 | | Release date: | 1999-06-14 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2.75 Å) | | Cite: | Structure of human erythrocyte catalase.
Acta Crystallogr.,Sect.D, 56, 2000
|
|
1TD2
 
 | | Crystal Structure of the PdxY Protein from Escherichia coli | | Descriptor: | 3-HYDROXY-5-(HYDROXYMETHYL)-2-METHYLISONICOTINALDEHYDE, Pyridoxamine kinase, SULFATE ION | | Authors: | Safo, M.K, Musayev, F.N, Hunt, S, di Salvo, M, Scarsdale, N, Schirch, V. | | Deposit date: | 2004-05-21 | | Release date: | 2004-07-13 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (2.22 Å) | | Cite: | Crystal structure of the PdxY Protein from Escherichia coli
J.Bacteriol., 186, 2004
|
|
1RV3
 
 | | E75L MUTANT OF RABBIT CYTOSOLIC SERINE HYDROXYMETHYLTRANSFERASE, COMPLEX WITH GLYCINE | | Descriptor: | GLYCINE, PHOSPHATE ION, PYRIDOXAL-5'-PHOSPHATE, ... | | Authors: | Szebenyi, D.M, Musayev, F.N, di Salvo, M.L, Safo, M.K, Schirch, V. | | Deposit date: | 2003-12-12 | | Release date: | 2004-06-15 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Serine Hydroxymethyltransferase: Role of Glu75 and Evidence that Serine Is Cleaved by a Retroaldol Mechanism.
Biochemistry, 43, 2004
|
|
3FHY
 
 | | Crystal structure of D235N mutant of human pyridoxal kinase | | Descriptor: | (4S)-2-METHYL-2,4-PENTANEDIOL, ADENOSINE-5'-TRIPHOSPHATE, MAGNESIUM ION, ... | | Authors: | Safo, M.K, Gandhi, A.K, Musayev, F.N, Ghatge, M, Di Salvo, M.L, Schirch, V. | | Deposit date: | 2008-12-10 | | Release date: | 2008-12-23 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Kinetic and structural studies of the role of the active site residue Asp235 of human pyridoxal kinase.
Biochem.Biophys.Res.Commun., 381, 2009
|
|
3GRR
 
 | |
3GRV
 
 | |
3FYD
 
 | |
3FYC
 
 | |
3HZZ
 
 | |
3GRU
 
 | |
3FHX
 
 | | Crystal structure of D235A mutant of human pyridoxal kinase | | Descriptor: | (4S)-2-METHYL-2,4-PENTANEDIOL, 3-HYDROXY-5-(HYDROXYMETHYL)-2-METHYLISONICOTINALDEHYDE, ADENOSINE-5'-TRIPHOSPHATE, ... | | Authors: | Safo, M.K, Gandhi, A.K, Musayev, F.N, Ghatge, M, Di Salvo, M.L, Schirch, V. | | Deposit date: | 2008-12-10 | | Release date: | 2008-12-23 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Kinetic and structural studies of the role of the active site residue Asp235 of human pyridoxal kinase.
Biochem.Biophys.Res.Commun., 381, 2009
|
|
3GRY
 
 | |
7UAX
 
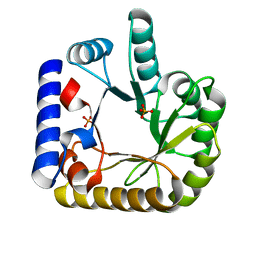 | | The crystal structure of the K36A/K38A double mutant of E. coli YGGS in complex with PLP | | Descriptor: | PHOSPHATE ION, Pyridoxal phosphate homeostasis protein | | Authors: | Donkor, A.K, Ghatge, M.S, Musayev, F.N, Safo, M.K. | | Deposit date: | 2022-03-14 | | Release date: | 2022-03-23 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (2.07 Å) | | Cite: | Characterization of the Escherichia coli pyridoxal 5'-phosphate homeostasis protein (YggS): Role of lysine residues in PLP binding and protein stability.
Protein Sci., 31, 2022
|
|
7U9H
 
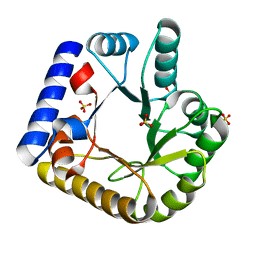 | | Crystal Structure of Escherichia coli apo Pyridoxal 5'-phosphate homeostasis protein (YGGS) | | Descriptor: | Pyridoxal phosphate homeostasis protein, SULFATE ION | | Authors: | Donkor, A.K, Ghatge, M.S, Musayev, F.N, Safo, M.K. | | Deposit date: | 2022-03-10 | | Release date: | 2022-03-23 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Characterization of the Escherichia coli pyridoxal 5'-phosphate homeostasis protein (YggS): Role of lysine residues in PLP binding and protein stability.
Protein Sci., 31, 2022
|
|
7UAU
 
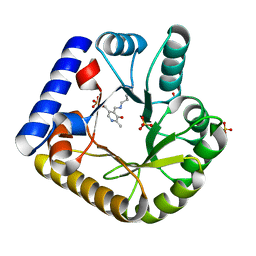 | | The crystal structure of the K137A mutant of E. coli YGGS in complex with PLP | | Descriptor: | PYRIDOXAL-5'-PHOSPHATE, Pyridoxal phosphate homeostasis protein, SULFATE ION | | Authors: | Donkor, A.K, Ghatge, M.S, Musayev, F.N, Safo, M.K. | | Deposit date: | 2022-03-14 | | Release date: | 2022-03-23 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Characterization of the Escherichia coli pyridoxal 5'-phosphate homeostasis protein (YggS): Role of lysine residues in PLP binding and protein stability.
Protein Sci., 31, 2022
|
|
