6FYB
 
 | | The crystal structure of EncM L144M mutant | | Descriptor: | FLAVIN-ADENINE DINUCLEOTIDE, Putative FAD-dependent oxygenase EncM | | Authors: | Saleem-Batcha, R, Teufel, R. | | Deposit date: | 2018-03-11 | | Release date: | 2018-05-02 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (1.882 Å) | | Cite: | Enzymatic control of dioxygen binding and functionalization of the flavin cofactor.
Proc. Natl. Acad. Sci. U.S.A., 115, 2018
|
|
6FYC
 
 | |
6FOW
 
 | |
2D09
 
 | |
2Q6K
 
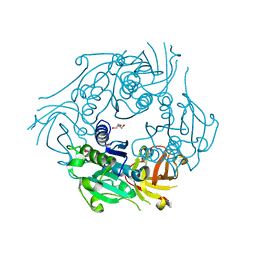 | | SalL with adenosine | | Descriptor: | ADENOSINE, DI(HYDROXYETHYL)ETHER, chlorinase | | Authors: | Pojer, F, Noel, J.P. | | Deposit date: | 2007-06-05 | | Release date: | 2007-12-11 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (1.55 Å) | | Cite: | Discovery and characterization of a marine bacterial SAM-dependent chlorinase
Nat.Chem.Biol., 4, 2008
|
|
2Q6O
 
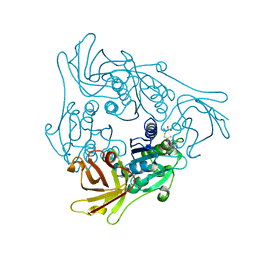 | | SalL-Y70T with SAM and Cl | | Descriptor: | CHLORIDE ION, Hypothetical Protein, S-ADENOSYLMETHIONINE | | Authors: | Noel, J.P, Pojer, F. | | Deposit date: | 2007-06-05 | | Release date: | 2007-12-11 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Discovery and characterization of a marine bacterial SAM-dependent chlorinase
Nat.Chem.Biol., 4, 2008
|
|
2Q6I
 
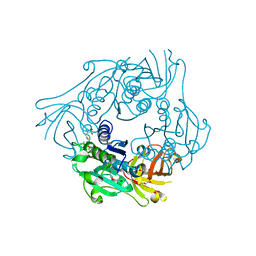 | | salL with ClDA and LMet | | Descriptor: | 5'-CHLORO-5'-DEOXYADENOSINE, DI(HYDROXYETHYL)ETHER, Hypothetical protein, ... | | Authors: | Noel, J.P, Pojer, F. | | Deposit date: | 2007-06-05 | | Release date: | 2007-12-11 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Discovery and characterization of a marine bacterial SAM-dependent chlorinase
Nat.Chem.Biol., 4, 2008
|
|
2Q6L
 
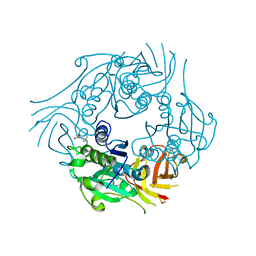 | | SalL double mutant Y70T/G131S with CLDA and L-MET | | Descriptor: | 5'-CHLORO-5'-DEOXYADENOSINE, Hypothetical protein, METHIONINE | | Authors: | Pojer, F, Noel, J.P. | | Deposit date: | 2007-06-05 | | Release date: | 2007-12-11 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (2.72 Å) | | Cite: | Discovery and characterization of a marine bacterial SAM-dependent chlorinase
Nat.Chem.Biol., 4, 2008
|
|
1T93
 
 | | Evidence for Multiple Substrate Recognition and Molecular Mechanism of C-C reaction by Cytochrome P450 CYP158A2 from Streptomyces Coelicolor A3(2) | | Descriptor: | FLAVIOLIN, PROTOPORPHYRIN IX CONTAINING FE, putative cytochrome P450 | | Authors: | Zhao, B, Sundaramoorthy, M, Waterman, M.R. | | Deposit date: | 2004-05-14 | | Release date: | 2005-01-25 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (1.62 Å) | | Cite: | Binding of Two Flaviolin Substrate Molecules, Oxidative Coupling, and Crystal Structure of Streptomyces coelicolor A3(2) Cytochrome P450 158A2.
J.Biol.Chem., 280, 2005
|
|
1S1F
 
 | | Crystal Structure of Streptomyces Coelicolor A3(2) CYP158A2 from antibiotic biosynthetic pathways | | Descriptor: | 4-PHENYL-1H-IMIDAZOLE, GLYCEROL, MALONIC ACID, ... | | Authors: | Zhao, B, Lamb, D.C, Lei, L, Sundaramoorthy, M, Podust, L.M, Waterman, M.R. | | Deposit date: | 2004-01-06 | | Release date: | 2005-01-11 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Binding of Two Flaviolin Substrate Molecules, Oxidative Coupling, and Crystal Structure of Streptomyces coelicolor A3(2) Cytochrome P450 158A2
J.Biol.Chem., 280, 2005
|
|
1SE6
 
 | | Crystal Structure of Streptomyces Coelicolor A3(2) CYP158A2 from antibiotic biosynthetic pathways | | Descriptor: | 2-(N-MORPHOLINO)-ETHANESULFONIC ACID, PROTOPORPHYRIN IX CONTAINING FE, SPERMINE (FULLY PROTONATED FORM), ... | | Authors: | Zhao, B, Lamb, D.C, Lei, L, Sundaramoorthy, M, Podust, L.M, Waterman, M.R. | | Deposit date: | 2004-02-16 | | Release date: | 2005-01-25 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (1.75 Å) | | Cite: | Binding of Two Flaviolin Substrate Molecules, Oxidative Coupling, and Crystal Structure of Streptomyces coelicolor A3(2) Cytochrome P450 158A2.
J.Biol.Chem., 280, 2005
|
|
