2VEE
 
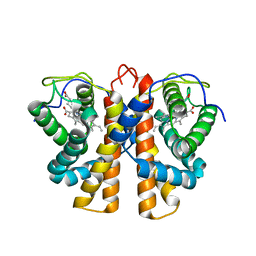 | | Structure of protoglobin from Methanosarcina acetivorans C2A | | Descriptor: | PROTOGLOBIN, PROTOPORPHYRIN IX CONTAINING FE | | Authors: | Nardini, M, Pesce, A, Thijs, L, Saito, J.A, Dewilde, S, Alam, M, Ascenzi, P, Coletta, M, Ciaccio, C, Moens, L, Bolognesi, M. | | Deposit date: | 2007-10-22 | | Release date: | 2008-01-22 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Archaeal Protoglobin Structure Indicates New Ligand Diffusion Paths and Modulation of Haem-Reactivity.
Embo Rep., 9, 2008
|
|
2VEB
 
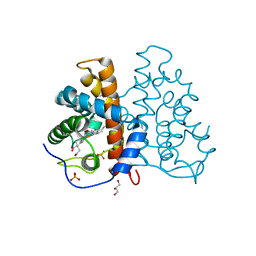 | | High resolution structure of protoglobin from Methanosarcina acetivorans C2A | | Descriptor: | GLYCEROL, OXYGEN MOLECULE, PHOSPHATE ION, ... | | Authors: | Nardini, M, Pesce, A, Thijs, L, Saito, J.A, Dewilde, S, Alam, M, Ascenzi, P, Coletta, M, Ciaccio, C, Moens, L, Bolognesi, M. | | Deposit date: | 2007-10-18 | | Release date: | 2008-01-22 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (1.3 Å) | | Cite: | Archaeal Protoglobin Structure Indicates New Ligand Diffusion Paths and Modulation of Haem-Reactivity.
Embo Rep., 9, 2008
|
|
7Z9F
 
 | | Human anionic trypsin after autoproteolysis at Arg122 | | Descriptor: | CALCIUM ION, Trypsin-2 | | Authors: | Nagel, F, Palm, G.J, Delcea, M, Lammers, M. | | Deposit date: | 2022-03-21 | | Release date: | 2022-06-29 | | Last modified: | 2024-05-01 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Structural Basis of the Pancreatitis-Associated Autoproteolytic Failsafe Mechanism in Human Anionic Trypsin.
J Inflamm Res, 15, 2022
|
|
7AC0
 
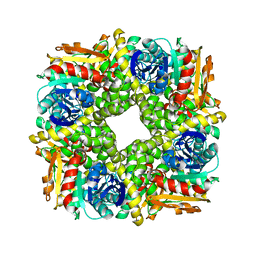 | | Epoxide hydrolase CorEH without ligand | | Descriptor: | Soluble epoxide hydrolase | | Authors: | Palm, G.J, Lammers, M, Berndt, L. | | Deposit date: | 2020-09-09 | | Release date: | 2021-07-21 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (2.177 Å) | | Cite: | Promiscuous Dehalogenase Activity of the Epoxide Hydrolase CorEH from Corynebacterium sp. C12
Acs Catalysis, 11, 2021
|
|
2W31
 
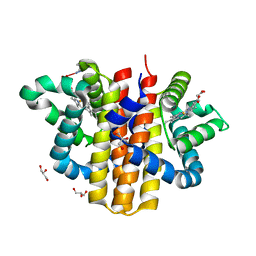 | | globin domain of Geobacter sulfurreducens globin-coupled sensor | | Descriptor: | GLOBIN, GLYCEROL, PROTOPORPHYRIN IX CONTAINING FE | | Authors: | Pesce, A, Thijs, L, Nardini, M, Desmet, F, Sisinni, L, Gourlay, L, Bolli, A, Coletta, M, Van Doorslaer, S, Wan, X, Alam, M, Ascenzi, P, Moens, L, Bolognesi, M, Dewilde, S. | | Deposit date: | 2008-11-05 | | Release date: | 2009-01-13 | | Last modified: | 2019-03-06 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Hise11 and Hisf8 Provide Bis-Histidyl Heme Hexa-Coordination in the Globin Domain of Geobacter Sulfurreducens Globin-Coupled Sensor.
J.Mol.Biol., 386, 2009
|
|
3NGO
 
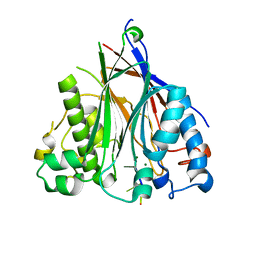 | | Crystal structure of the human CNOT6L nuclease domain in complex with poly(A) DNA | | Descriptor: | 5'-D(*AP*AP*AP*A)-3', CCR4-NOT transcription complex subunit 6-like, MAGNESIUM ION | | Authors: | Wang, H, Morita, M, Yang, W, Bartlam, M, Yamamoto, T, Rao, Z. | | Deposit date: | 2010-06-12 | | Release date: | 2010-07-28 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Crystal structure of the human CNOT6L nuclease domain reveals strict poly(A) substrate specificity.
Embo J., 2010
|
|
3NGQ
 
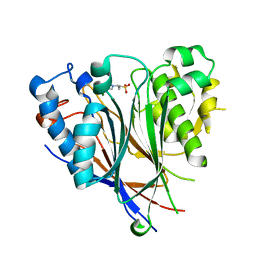 | | Crystal structure of the human CNOT6L nuclease domain | | Descriptor: | 3-PYRIDINIUM-1-YLPROPANE-1-SULFONATE, CCR4-NOT transcription complex subunit 6-like, MAGNESIUM ION | | Authors: | Wang, H, Morita, M, Yang, W, Bartlam, M, Yamamoto, T, Rao, Z. | | Deposit date: | 2010-06-13 | | Release date: | 2010-07-28 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Crystal structure of the human CNOT6L nuclease domain reveals strict poly(A) substrate specificity.
Embo J., 2010
|
|
3NGN
 
 | | Crystal structure of the human CNOT6L nuclease domain in complex with AMP | | Descriptor: | ADENOSINE MONOPHOSPHATE, CCR4-NOT transcription complex subunit 6-like | | Authors: | Wang, H, Morita, M, Yang, W, Bartlam, M, Yamamoto, T, Rao, Z. | | Deposit date: | 2010-06-12 | | Release date: | 2010-07-28 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Crystal structure of the human CNOT6L nuclease domain reveals strict poly(A) substrate specificity.
Embo J., 2010
|
|
8K8K
 
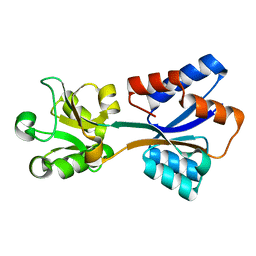 | | Structure of Klebsiella pneumonia ModA | | Descriptor: | Molybdate transporter periplasmic protein | | Authors: | Zhao, Q, Bartlam, M. | | Deposit date: | 2023-07-31 | | Release date: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (1.99 Å) | | Cite: | Structural analysis of molybdate binding protein ModA from Klebsiella pneumoniae.
Biochem.Biophys.Res.Commun., 681, 2023
|
|
8K8L
 
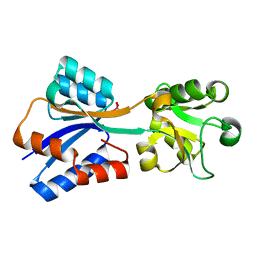 | |
8K7N
 
 | | Structure of ferric-binding protein KfuA | | Descriptor: | FE (III) ION, Ferric iron ABC transporter, iron-binding protein | | Authors: | Zhao, Q, Bartlam, M. | | Deposit date: | 2023-07-26 | | Release date: | 2024-07-31 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Structure of Klebsiella pneumoniae ferric-binding protein KfuA
To be published
|
|
1JS1
 
 | | Crystal Structure of a new transcarbamylase from the anaerobic bacterium Bacteroides fragilis at 2.0 A resolution | | Descriptor: | PHOSPHATE ION, Transcarbamylase | | Authors: | Shi, D, Gallegos, R, DePonte III, J, Morizono, H, Yu, X, Allewell, N.M, Malamy, M, Tuchman, M. | | Deposit date: | 2001-08-15 | | Release date: | 2002-07-17 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Crystal structure of a transcarbamylase-like protein from the anaerobic bacterium Bacteroides fragilis at 2.0 A resolution.
J.Mol.Biol., 320, 2002
|
|
3EBJ
 
 | | Crystal structure of an avian influenza virus protein | | Descriptor: | MAGNESIUM ION, Polymerase acidic protein | | Authors: | Yuan, P, Bartlam, M, Lou, Z, Chen, S, Rao, Z, Liu, Y. | | Deposit date: | 2008-08-27 | | Release date: | 2009-02-10 | | Last modified: | 2024-05-29 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Crystal structure of an avian influenza polymerase PA(N) reveals an endonuclease active site
Nature, 458, 2009
|
|
1UJ1
 
 | | Crystal structure of SARS Coronavirus Main Proteinase (3CLpro) | | Descriptor: | 3C-like proteinase | | Authors: | Yang, H, Yang, M, Liu, Y, Bartlam, M, Ding, Y, Lou, Z, Sun, L, Zhou, Z, Ye, S, Anand, K, Pang, H, Gao, G.F, Hilgenfeld, R, Rao, Z. | | Deposit date: | 2003-07-25 | | Release date: | 2003-11-18 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | The crystal structures of severe acute respiratory syndrome virus main protease and its complex with an inhibitor
Proc.Natl.Acad.Sci.USA, 100, 2003
|
|
1UK2
 
 | | Crystal structure of SARS Coronavirus Main Proteinase (3CLpro) At pH8.0 | | Descriptor: | 3C-LIKE PROTEINASE | | Authors: | Yang, H, Yang, M, Liu, Y, Bartlam, M, Ding, Y, Lou, Z, Sun, L, Zhou, Z, Ye, S, Anand, K, Pang, H, Gao, G.F, Hilgenfeld, R, Rao, Z. | | Deposit date: | 2003-08-14 | | Release date: | 2003-11-18 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | The crystal structures of severe acute respiratory syndrome virus main protease and its complex with an inhibitor
Proc.Natl.Acad.Sci.USA, 100, 2003
|
|
1UK3
 
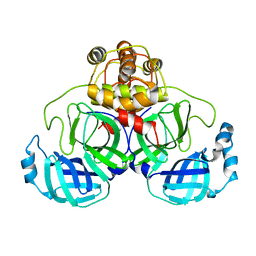 | | Crystal structure of SARS Coronavirus Main Proteinase (3CLpro) At pH7.6 | | Descriptor: | 3C-like proteinase | | Authors: | Yang, H, Yang, M, Liu, Y, Bartlam, M, Ding, Y, Lou, Z, Sun, L, Zhou, Z, Ye, S, Anand, K, Pang, H, Gao, G.F, Hilgenfeld, R, Rao, Z. | | Deposit date: | 2003-08-14 | | Release date: | 2003-11-18 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | The crystal structures of severe acute respiratory syndrome virus main protease and its complex with an inhibitor
Proc.Natl.Acad.Sci.USA, 100, 2003
|
|
1UK4
 
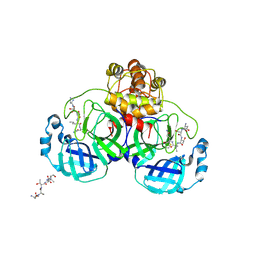 | | Crystal structure of SARS Coronavirus Main Proteinase (3CLpro) Complexed With An Inhibitor | | Descriptor: | 3C-like proteinase nsp5, 5-mer peptide of inhibitor | | Authors: | Yang, H, Yang, M, Liu, Y, Bartlam, M, Ding, Y, Lou, Z, Sun, L, Zhou, Z, Ye, S, Anand, K, Pang, H, Gao, G.F, Hilgenfeld, R, Rao, Z. | | Deposit date: | 2003-08-14 | | Release date: | 2003-11-18 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | The crystal structures of severe acute respiratory syndrome virus main protease and its complex with an inhibitor
Proc.Natl.Acad.Sci.USA, 100, 2003
|
|
7BVD
 
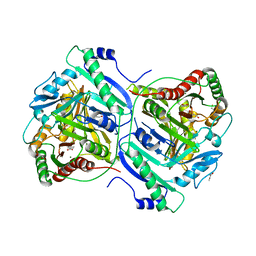 | | Anthranilate synthase component I (TrpE)[Mycolicibacterium smegmatis] | | Descriptor: | Anthranilate synthase component 1, BENZOIC ACID, GLYCEROL, ... | | Authors: | Chen, Y, Che, S, Zhang, Q, Bartlam, M. | | Deposit date: | 2020-04-10 | | Release date: | 2020-05-06 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Structure of subunit I of the anthranilate synthase complex of Mycolicibacterium smegmatis
Biochem.Biophys.Res.Commun., 527, 2020
|
|
4R0S
 
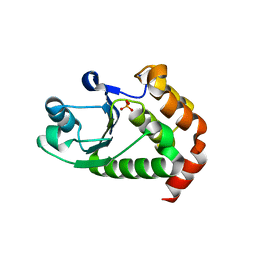 | | Crystal structure of P. aeruginosa TpbA | | Descriptor: | GLYCEROL, PHOSPHATE ION, Protein tyrosine phosphatase TpbA | | Authors: | Xu, K, Li, S, Wang, Y, Bartlam, M. | | Deposit date: | 2014-08-01 | | Release date: | 2015-05-06 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (2.03 Å) | | Cite: | Structural and Biochemical Analysis of Tyrosine Phosphatase Related to Biofilm Formation A (TpbA) from the Opportunistic Pathogen Pseudomonas aeruginosa PAO1
PLoS ONE, 10, 2015
|
|
7D54
 
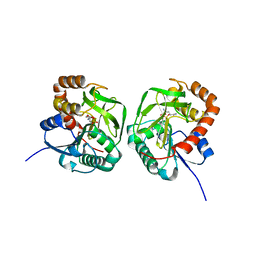 | | Crstal structure MsGATase with Gln | | Descriptor: | GLUTAMINE, Glutamine amidotransferase class-I | | Authors: | Chen, Y, Zhang, Q, Bartlam, M. | | Deposit date: | 2020-09-24 | | Release date: | 2021-10-06 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | Structure and mechanism of the gamma-glutamyl-gamma-aminobutyrate hydrolase SpuA from Pseudomonas aeruginosa.
Acta Crystallogr D Struct Biol, 77, 2021
|
|
7D4R
 
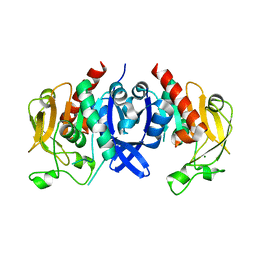 | | SpuA native structure | | Descriptor: | MAGNESIUM ION, Probable glutamine amidotransferase | | Authors: | Chen, Y, Zhang, Q, Bartlam, M. | | Deposit date: | 2020-09-24 | | Release date: | 2021-10-06 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Structure and mechanism of the gamma-glutamyl-gamma-aminobutyrate hydrolase SpuA from Pseudomonas aeruginosa.
Acta Crystallogr D Struct Biol, 77, 2021
|
|
7D53
 
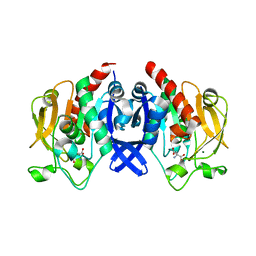 | | SpuA mutant - H221N with Glu | | Descriptor: | GLUTAMIC ACID, MAGNESIUM ION, Probable glutamine amidotransferase | | Authors: | Chen, Y, Zhang, Q, Bartlam, M. | | Deposit date: | 2020-09-24 | | Release date: | 2021-10-06 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Structure and mechanism of the gamma-glutamyl-gamma-aminobutyrate hydrolase SpuA from Pseudomonas aeruginosa.
Acta Crystallogr D Struct Biol, 77, 2021
|
|
7D50
 
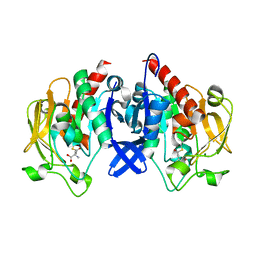 | | SpuA mutant - H221N with glutamyl-thioester | | Descriptor: | MAGNESIUM ION, Probable glutamine amidotransferase | | Authors: | Chen, Y, Zhang, Q, Bartlam, M. | | Deposit date: | 2020-09-24 | | Release date: | 2021-10-06 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (1.77 Å) | | Cite: | Structure and mechanism of the gamma-glutamyl-gamma-aminobutyrate hydrolase SpuA from Pseudomonas aeruginosa.
Acta Crystallogr D Struct Biol, 77, 2021
|
|
7D9F
 
 | | SpdH Spermidine dehydrogenase SeMet Structure | | Descriptor: | FLAVIN-ADENINE DINUCLEOTIDE, GLYCEROL, PROTOPORPHYRIN IX CONTAINING FE, ... | | Authors: | Che, S, Zhang, Q, Bartlam, M. | | Deposit date: | 2020-10-13 | | Release date: | 2021-11-17 | | Last modified: | 2022-04-20 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | Structure of Pseudomonas aeruginosa spermidine dehydrogenase: a polyamine oxidase with a novel heme-binding fold.
Febs J., 289, 2022
|
|
7D9J
 
 | | SpdH Spermidine dehydrogenase Y443A mutant | | Descriptor: | FLAVIN-ADENINE DINUCLEOTIDE, GLYCEROL, PROTOPORPHYRIN IX CONTAINING FE, ... | | Authors: | Che, S, Zhang, Q, Bartlam, M. | | Deposit date: | 2020-10-13 | | Release date: | 2021-11-17 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.19 Å) | | Cite: | Structure of Pseudomonas aeruginosa spermidine dehydrogenase: a polyamine oxidase with a novel heme-binding fold.
Febs J., 289, 2022
|
|
