3O6C
 
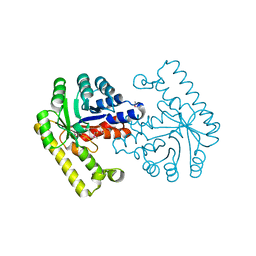 | | Pyridoxal phosphate biosynthetic protein PdxJ from Campylobacter jejuni | | Descriptor: | PHOSPHATE ION, Pyridoxine 5'-phosphate synthase | | Authors: | Osipiuk, J, Zhou, M, Kwon, K, Anderson, W.F, Joachimiak, A, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2010-07-28 | | Release date: | 2010-08-11 | | Last modified: | 2017-11-08 | | Method: | X-RAY DIFFRACTION (1.87 Å) | | Cite: | Pyridoxal phosphate biosynthetic protein PdxJ from Campylobacter jejuni.
To be Published
|
|
4EGR
 
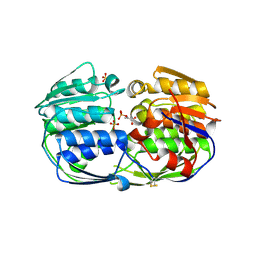 | | 2.50 angstrom resolution structure of 3-phosphoshikimate 1-carboxyvinyltransferase (AroA) from Coxiella burnetii in complex with phosphoenolpyruvate | | Descriptor: | 3-phosphoshikimate 1-carboxyvinyltransferase, PHOSPHOENOLPYRUVATE, SULFATE ION | | Authors: | Krishna, S.N, Light, S.H, Minasov, G, Shuvalova, L, Kwon, K, Anderson, W.F, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2012-03-31 | | Release date: | 2012-04-11 | | Last modified: | 2024-10-09 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | 2.50 angstrom resolution structure of 3-phosphoshikimate 1-carboxyvinyltransferase (AroA) from Coxiella burnetii in complex with phosphoenolpyruvate
TO BE PUBLISHED
|
|
4ECD
 
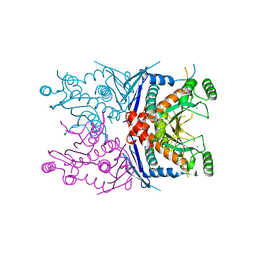 | | 2.5 Angstrom Resolution Crystal Structure of Bifidobacterium longum Chorismate Synthase | | Descriptor: | CHLORIDE ION, Chorismate synthase | | Authors: | Light, S.H, Minasov, G, Krishna, S.N, Shuvalova, L, Kwon, K, Lavie, A, Anderson, W.F, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2012-03-26 | | Release date: | 2012-04-04 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | 2.5 Angstrom Resolution Crystal Structure of Bifidobacterium longum Chorismate Synthase
TO BE PUBLISHED
|
|
3NUA
 
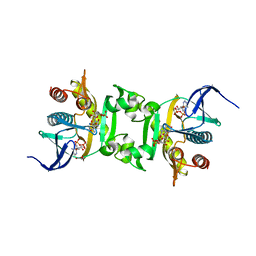 | | Crystal Structure of Phosphoribosylaminoimidazole-Succinocarboxamide Synthase from Clostridium perfringens | | Descriptor: | ADENOSINE MONOPHOSPHATE, ADENOSINE-5'-DIPHOSPHATE, CITRIC ACID, ... | | Authors: | Kim, Y, Mulligan, R, Kwon, K, Anderson, W.F, Joachimiak, A, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2010-07-06 | | Release date: | 2010-08-25 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (1.4 Å) | | Cite: | Crystal Structure of Phosphoribosylaminoimidazole-Succinocarboxamide Synthase from Clostridium perfringens
To be Published
|
|
4E6Y
 
 | | Type II citrate synthase from Vibrio vulnificus. | | Descriptor: | Citrate synthase, FORMIC ACID | | Authors: | Osipiuk, J, Gu, M, Kwon, K, Anderson, W.F, Joachimiak, A, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2012-03-16 | | Release date: | 2012-03-28 | | Last modified: | 2023-12-06 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Type II citrate synthase from Vibrio vulnificus.
To be Published
|
|
3OJC
 
 | | Crystal structure of a putative Asp/Glu Racemase from Yersinia pestis | | Descriptor: | CALCIUM ION, HEXANE-1,6-DIOL, Putative aspartate/glutamate racemase | | Authors: | Anderson, S.M, Wawrzak, Z, Kudritska, M, Kwon, K, Anderson, W.F, Savchenko, A, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2010-08-21 | | Release date: | 2010-09-15 | | Last modified: | 2017-11-08 | | Method: | X-RAY DIFFRACTION (1.75 Å) | | Cite: | Crystal structure of a putative Asp/Glu Racemase from Yersinia pestis
To be Published
|
|
3OO2
 
 | | 2.37 Angstrom resolution crystal structure of an alanine racemase (alr) from Staphylococcus aureus subsp. aureus COL | | Descriptor: | Alanine racemase 1, BETA-MERCAPTOETHANOL, PHOSPHATE ION, ... | | Authors: | Halavaty, A.S, Shuvalova, L, Minasov, G, Winsor, J, Dubrovska, I, Kwon, K, Anderson, W.F, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2010-08-30 | | Release date: | 2010-10-06 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (2.37 Å) | | Cite: | 2.37 Angstrom resolution crystal structure of an alanine racemase (alr) from Staphylococcus aureus subsp. aureus COL
TO BE PUBLISHED
|
|
4E0B
 
 | | 2.17 Angstrom resolution crystal structure of malate dehydrogenase from Vibrio vulnificus CMCP6 | | Descriptor: | ACETATE ION, Malate dehydrogenase | | Authors: | Halavaty, A.S, Wawrzak, Z, Onopriyenko, O, Kwon, K, Anderson, W.F, Savchenko, A, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2012-03-02 | | Release date: | 2012-03-21 | | Last modified: | 2023-12-06 | | Method: | X-RAY DIFFRACTION (2.17 Å) | | Cite: | 2.17 Angstrom resolution crystal structure of malate dehydrogenase from Vibrio vulnificus CMCP6
To be Published
|
|
3M5V
 
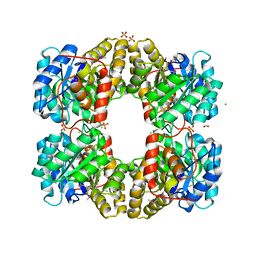 | | Crystal Structure of Dihydrodipicolinate Synthase from Campylobacter jejuni | | Descriptor: | CHLORIDE ION, DI(HYDROXYETHYL)ETHER, Dihydrodipicolinate synthase, ... | | Authors: | Kim, Y, Zhou, M, Kwon, K, Anderson, W.F, Joachimiak, A, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2010-03-13 | | Release date: | 2010-04-28 | | Last modified: | 2012-02-29 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Crystal Structure of Dihydrodipicolinate Synthase from Campylobacter jejuni
To be Published
|
|
3NAV
 
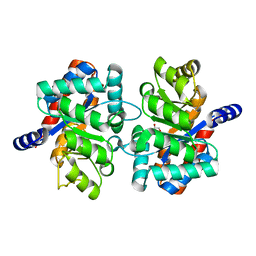 | | Crystal structure of an alpha subunit of tryptophan synthase from Vibrio cholerae O1 biovar El Tor str. N16961 | | Descriptor: | 1,2-ETHANEDIOL, Tryptophan synthase alpha chain | | Authors: | Nocek, B, Makowska-Grzyska, M, Kwon, K, Anderson, A, Joachimiak, A, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2010-06-02 | | Release date: | 2010-06-30 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Crystal structure of an alpha subunit of tryptophan synthase from Vibrio cholerae O1 biovar El Tor str. N16961
To be Published
|
|
3OWA
 
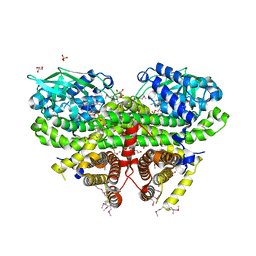 | | Crystal Structure of Acyl-CoA Dehydrogenase complexed with FAD from Bacillus anthracis | | Descriptor: | Acyl-CoA dehydrogenase, DI(HYDROXYETHYL)ETHER, FLAVIN-ADENINE DINUCLEOTIDE, ... | | Authors: | Kim, Y, Maltseva, N, Kwon, K, Anderson, W.F, Joachimiak, A, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2010-09-17 | | Release date: | 2010-10-13 | | Last modified: | 2023-12-06 | | Method: | X-RAY DIFFRACTION (1.97 Å) | | Cite: | Crystal Structure of Acyl-CoA Dehydrogenase complexed with FAD from Bacillus anthracis
To be Published
|
|
4HDE
 
 | | The crystal structure of a SCO1/SenC family lipoprotein from Bacillus anthracis str. Ames | | Descriptor: | SCO1/SenC family lipoprotein | | Authors: | Tan, K, Zhou, M, Kwon, K, Anderson, W.F, Joachimiak, A, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2012-10-02 | | Release date: | 2012-10-24 | | Last modified: | 2024-10-09 | | Method: | X-RAY DIFFRACTION (1.317 Å) | | Cite: | The crystal structure of a SCO1/SenC family lipoprotein from Bacillus anthracis str. Ames
To be Published
|
|
4DU6
 
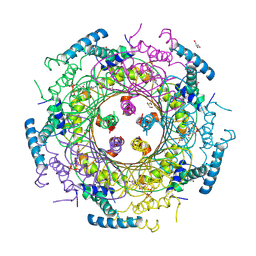 | | Crystal structure of GTP cyclohydrolase I from Yersinia pestis complexed with GTP | | Descriptor: | 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, CALCIUM ION, DI(HYDROXYETHYL)ETHER, ... | | Authors: | Maltseva, N, Kim, Y, Kwon, K, Anderson, W.F, Joachimiak, A, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2012-02-21 | | Release date: | 2012-03-21 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (2.106 Å) | | Cite: | Crystal structure of GTP cyclohydrolase I from Yersinia pestis complexed with GTP
To be Published
|
|
3N2L
 
 | | 2.1 Angstrom resolution crystal structure of an Orotate Phosphoribosyltransferase (pyrE) from Vibrio cholerae O1 biovar eltor str. N16961 | | Descriptor: | CHLORIDE ION, Orotate phosphoribosyltransferase | | Authors: | Halavaty, A.S, Minasov, G, Shuvalova, L, Dubrovska, I, Winsor, J, Kwon, K, Anderson, W.F, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2010-05-18 | | Release date: | 2010-06-02 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | 2.1 Angstrom resolution crystal structure of an Orotate Phosphoribosyltransferase
(pyrE) from Vibrio cholerae O1 biovar eltor str. N16961
To be Published
|
|
3PGY
 
 | | Serine hydroxymethyltransferase from Staphylococcus aureus, S95P mutant. | | Descriptor: | 1,2-ETHANEDIOL, CITRIC ACID, GLYCINE, ... | | Authors: | Osipiuk, J, Makowska-Grzyska, M, Kwon, K, Anderson, W.F, Joachimiak, A, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2010-11-02 | | Release date: | 2010-11-17 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (1.92 Å) | | Cite: | Serine hydroxymethyltransferase from Staphylococcus aureus, S95P mutant
To be Published
|
|
3Q10
 
 | | Pantoate-beta-alanine ligase from Yersinia pestis | | Descriptor: | ADENOSINE MONOPHOSPHATE, CHLORIDE ION, GLYCEROL, ... | | Authors: | Osipiuk, J, Maltseva, N, Kwon, K, Anderson, W.F, Joachimiak, A, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2010-12-16 | | Release date: | 2011-02-02 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (1.83 Å) | | Cite: | Pantoate-beta-alanine ligase from Yersinia pestis.
To be Published
|
|
4HN3
 
 | | The crystal structure of a sex pheromone precursor (lmo1757) from Listeria monocytogenes EGD-e | | Descriptor: | BETA-MERCAPTOETHANOL, DI(HYDROXYETHYL)ETHER, GLYCEROL, ... | | Authors: | Tan, K, Makowska-Grzyska, M, Kwon, K, Anderson, W.F, Joachimiak, A, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2012-10-18 | | Release date: | 2012-10-31 | | Method: | X-RAY DIFFRACTION (2.047 Å) | | Cite: | The crystal structure of a sex pheromone precursor (lmo1757) from Listeria monocytogenes EGD-e
To be Published
|
|
3Q12
 
 | | Pantoate-beta-alanine ligase from Yersinia pestis in complex with pantoate. | | Descriptor: | CHLORIDE ION, PANTOATE, Pantoate--beta-alanine ligase | | Authors: | Osipiuk, J, Maltseva, N, Kwon, K, Anderson, W.F, Joachimiak, A, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2010-12-16 | | Release date: | 2011-02-02 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (1.58 Å) | | Cite: | Pantoate-beta-alanine ligase from Yersinia pestis.
To be Published
|
|
4FK1
 
 | | Crystal Structure of Putative Thioredoxin Reductase TrxB from Bacillus anthracis | | Descriptor: | FLAVIN-ADENINE DINUCLEOTIDE, GLYCEROL, MAGNESIUM ION, ... | | Authors: | Maltseva, N, Kim, Y, Kwon, K, Anderson, W.F, Joachimiak, A, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2012-06-12 | | Release date: | 2012-08-08 | | Last modified: | 2023-12-06 | | Method: | X-RAY DIFFRACTION (2.404 Å) | | Cite: | Crystal Structure of Putative Thioredoxin Reductase TrxB from Bacillus anthracis
To be Published
|
|
5UBU
 
 | | 2.75 Angstrom Resolution Crystal Structure of Acetamidase from Yersinia enterocolitica. | | Descriptor: | Putative acetamidase/formamidase, SODIUM ION | | Authors: | Minasov, G, Shuvalova, L, Flores, K, Dubrovska, I, Grimshaw, S, Kwon, K, Anderson, W.F, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2016-12-21 | | Release date: | 2017-01-04 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (2.75 Å) | | Cite: | 2.75 Angstrom Resolution Crystal Structure of Acetamidase from Yersinia enterocolitica.
To Be Published
|
|
5TW9
 
 | | 1.50 Angstrom Crystal Structure of C-terminal Fragment (residues 322-384) of Iron Uptake System Component EfeO from Yersinia pestis. | | Descriptor: | 1,2-ETHANEDIOL, IODIDE ION, Iron uptake system component EfeO | | Authors: | Minasov, G, Shuvalova, L, Flores, K, Dubrovska, I, Grimshaw, S, Kwon, K, Anderson, W.F, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2016-11-11 | | Release date: | 2016-11-30 | | Last modified: | 2024-03-06 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | 1.50 Angstrom Crystal Structure of C-terminal Fragment (residues 322-384) of Iron Uptake System Component EfeO from Yersinia pestis.
To Be Published
|
|
5UH0
 
 | | 1.95 Angstrom Resolution Crystal Structure of Fragment (35-274) of Membrane-bound Lytic Murein Transglycosylase F from Yersinia pestis. | | Descriptor: | CHLORIDE ION, Membrane-bound lytic murein transglycosylase F | | Authors: | Minasov, G, Shuvalova, L, Flores, K, Kiryukhina, O, Grimshaw, S, Kwon, K, Anderson, W.F, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2017-01-10 | | Release date: | 2017-01-25 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | 1.95 Angstrom Resolution Crystal Structure of Fragment (35-274) of Membrane-bound Lytic Murein Transglycosylase F from Yersinia pestis.
To Be Published
|
|
5U9C
 
 | | 1.9 Angstrom Resolution Crystal Structure of dTDP-4-dehydrorhamnose Reductase from Yersinia enterocolitica | | Descriptor: | 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, CHLORIDE ION, CITRIC ACID, ... | | Authors: | Minasov, G, Shuvalova, L, Flores, K, Dubrovska, I, Olphie, A, Grimshaw, S, Kwon, K, Anderson, W.F, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2016-12-15 | | Release date: | 2016-12-28 | | Last modified: | 2020-07-29 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | 1.9 Angstrom Resolution Crystal Structure of dTDP-4-dehydrorhamnose Reductase from Yersinia enterocolitica.
To Be Published
|
|
5VGM
 
 | | Crystal structure of dihydroorotase pyrC from Vibrio cholerae in complex with zinc at 1.95 A resolution. | | Descriptor: | ACETATE ION, CHLORIDE ION, Dihydroorotase, ... | | Authors: | Lipowska, J, Shabalin, I.G, Miks, C.D, Winsor, J, Cooper, D.R, Shuvalova, L, Kwon, K, Lewinski, K, Anderson, W.F, Minor, W, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2017-04-11 | | Release date: | 2017-04-26 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | Pyrimidine biosynthesis in pathogens - Structures and analysis of dihydroorotases from Yersinia pestis and Vibrio cholerae.
Int.J.Biol.Macromol., 136, 2019
|
|
4PZ0
 
 | | The crystal structure of a solute binding protein from Bacillus anthracis str. Ames in complex with quorum-sensing signal autoinducer-2 (AI-2) | | Descriptor: | (2R,4S)-2-methyl-2,3,3,4-tetrahydroxytetrahydrofuran, 1,2-ETHANEDIOL, CHLORIDE ION, ... | | Authors: | Tan, K, Gu, M, Kwon, K, Anderson, W.F, Joachimiak, A, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2014-03-28 | | Release date: | 2014-04-09 | | Last modified: | 2020-07-29 | | Method: | X-RAY DIFFRACTION (1.25 Å) | | Cite: | The crystal structure of a solute binding protein from Bacillus anthracis str. Ames in complex with quorum-sensing signal autoinducer-2 (AI-2).
To be Published
|
|
