5H03
 
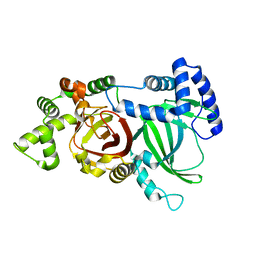 | | Crystal structure of an ADP-ribosylating toxin BECa from C. perfringens | | Descriptor: | Binary enterotoxin of Clostridium perfringens component a | | Authors: | Kawahara, K, Yonogi, S, Munetomo, R, Oki, H, Yoshida, T, Ohkubo, T, Kumeda, Y, Matsuda, S, Kodama, T, Iida, T, Nakamura, S. | | Deposit date: | 2016-10-03 | | Release date: | 2016-11-02 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (1.89 Å) | | Cite: | Crystal structure of the ADP-ribosylating component of BEC, the binary enterotoxin of Clostridium perfringens.
Biochem.Biophys.Res.Commun., 480, 2016
|
|
1EHD
 
 | |
1EHH
 
 | |
3T5W
 
 | | 2ME modified human SOD1 | | Descriptor: | COPPER (I) ION, SULFATE ION, Superoxide dismutase [Cu-Zn], ... | | Authors: | Ihara, K, Yamaguchi, Y, Torigoe, H, Wakatsuki, S, Taniguchi, N, Suzuki, K, Fujiwara, N. | | Deposit date: | 2011-07-28 | | Release date: | 2012-08-01 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Structural switching of Cu,Zn-superoxide dismutases at loop VI: insights from the crystal structure of 2-mercaptoethanol-modified enzyme
Biosci.Rep., 32, 2012
|
|
1IQB
 
 | |
1JSE
 
 | |
1JSF
 
 | |
2D4K
 
 | |
2D4I
 
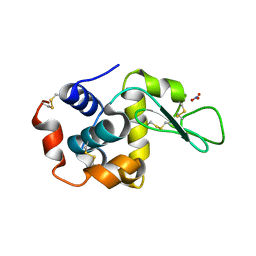 | |
2D4J
 
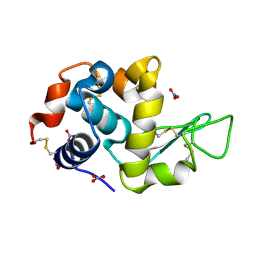 | |
3VTO
 
 | | The crystal structure of the C-terminal domain of Mu phage central spike | | Descriptor: | CALCIUM ION, CHLORIDE ION, FE (III) ION, ... | | Authors: | Harada, K, Yamashita, E, Nakagawa, A, Takeda, S. | | Deposit date: | 2012-06-01 | | Release date: | 2013-02-06 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (1.44 Å) | | Cite: | Crystal structure of the C-terminal domain of Mu phage central spike and functions of bound calcium ion
Biochim.Biophys.Acta, 1834, 2013
|
|
3VTN
 
 | | The crystal structure of the C-terminal domain of Mu phage central spike - Pt derivative for MAD | | Descriptor: | FE (III) ION, PLATINUM (II) ION, Protein gp45 | | Authors: | Harada, K, Yamashita, E, Nakagawa, A, Takeda, S. | | Deposit date: | 2012-06-01 | | Release date: | 2013-02-06 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (1.75 Å) | | Cite: | Crystal structure of the C-terminal domain of Mu phage central spike and functions of bound calcium ion
Biochim.Biophys.Acta, 1834, 2013
|
|
3WJ1
 
 | | Crystal structure of SSHESTI | | Descriptor: | Carboxylesterase, octyl beta-D-glucopyranoside | | Authors: | Ohara, K, Unno, H, Oshima, Y, Furukawa, K, Fujino, N, Hirooka, K, Hemmi, H, Takahashi, S, Nishino, T, Kusunoki, M, Nakayama, T. | | Deposit date: | 2013-10-03 | | Release date: | 2014-07-30 | | Last modified: | 2020-07-29 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Structural insights into the low pH adaptation of a unique carboxylesterase from Ferroplasma: altering the pH optima of two carboxylesterases.
J.Biol.Chem., 289, 2014
|
|
3WJ2
 
 | | Crystal structure of ESTFA (FE-lacking apo form) | | Descriptor: | Carboxylesterase | | Authors: | Ohara, K, Unno, H, Oshima, Y, Furukawa, K, Fujino, N, Hirooka, K, Hemmi, H, Takahashi, S, Nishino, T, Kusunoki, M, Nakayama, T. | | Deposit date: | 2013-10-03 | | Release date: | 2014-07-30 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (1.61 Å) | | Cite: | Structural insights into the low pH adaptation of a unique carboxylesterase from Ferroplasma: altering the pH optima of two carboxylesterases.
J.Biol.Chem., 289, 2014
|
|
2Z12
 
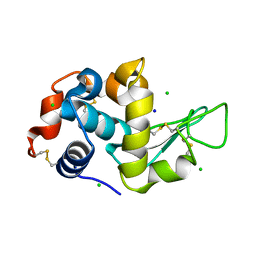 | |
2Z19
 
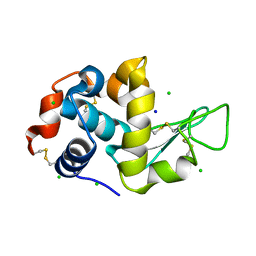 | |
2Z18
 
 | |
8W7D
 
 | | Crystal structure of EcPPAT-FR901483 complex | | Descriptor: | Amidophosphoribosyltransferase, [(1S,3S,6S,7S,8R,9S)-6-[(4-methoxyphenyl)methyl]-3-(methylamino)-7-oxidanyl-5-azatricyclo[6.3.1.0^1,5]dodecan-9-yl] dihydrogen phosphate | | Authors: | Hara, K, Hashimoto, H, Nakahara, M, Sato, M, Watanabe, K. | | Deposit date: | 2023-08-30 | | Release date: | 2023-12-13 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (2.81 Å) | | Cite: | Uncommon Arrangement of Self-resistance Allows Biosynthesis of de novo Purine Biosynthesis Inhibitor that Acts as an Immunosuppressor.
J.Am.Chem.Soc., 145, 2023
|
|
8YHR
 
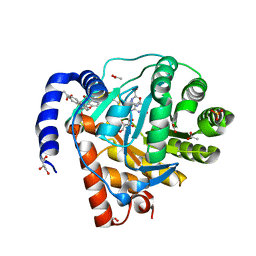 | | DHODH in complex with furocoumavirin | | Descriptor: | 4-methyl-8-[(S)-oxidanyl(phenyl)methyl]-9-phenyl-furo[2,3-h]chromen-2-one, ACETATE ION, Dihydroorotate dehydrogenase (quinone), ... | | Authors: | Hara, K, Okumura, H, Nakahara, M, Sato, M, Hashimoto, H, Osada, H, Watanabe, K. | | Deposit date: | 2024-02-28 | | Release date: | 2024-05-22 | | Last modified: | 2024-06-05 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Structural and Functional Analyses of Inhibition of Human Dihydroorotate Dehydrogenase by Antiviral Furocoumavirin.
Biochemistry, 63, 2024
|
|
8Y2S
 
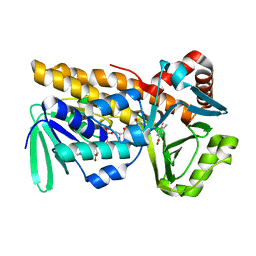 | | P-hydroxybenzoate hydroxylase complexed with 4-hydroxy-3-methylbenzoic acid | | Descriptor: | 3-methyl-4-oxidanyl-benzoic acid, 4-hydroxybenzoate 3-monooxygenase, FLAVIN-ADENINE DINUCLEOTIDE | | Authors: | Hara, K, Hashimoto, H, Matsushita, T, Kishimoto, S, Watanabe, K. | | Deposit date: | 2024-01-27 | | Release date: | 2024-05-01 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Functional Enhancement of Flavin-Containing Monooxygenase through Machine Learning Methodology
Acs Catalysis, 14, 2024
|
|
7BR0
 
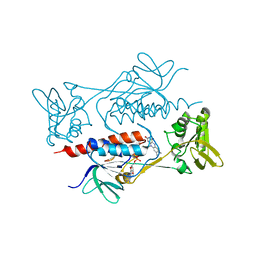 | | Crystal structure of AclR, a thioredoxin oxidoreductase fold protein carrying the CXXH catalytic motif | | Descriptor: | FLAVIN-ADENINE DINUCLEOTIDE, Pyr_redox_2 domain-containing protein | | Authors: | Hara, K, Hashimoto, H, Maeda, N, Watanabe, K, Hertweck, C, Tsunematsu, Y. | | Deposit date: | 2020-03-26 | | Release date: | 2020-12-23 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.003 Å) | | Cite: | Specialized Flavoprotein Promotes Sulfur Migration and Spiroaminal Formation in Aspirochlorine Biosynthesis.
J.Am.Chem.Soc., 143, 2021
|
|
5YY2
 
 | | Crystal structure of AsqI with Zn | | Descriptor: | Uncharacterized protein AsqI, ZINC ION | | Authors: | Hara, K, Hashimoto, H, Kishimoto, S, Watanabe, K. | | Deposit date: | 2017-12-07 | | Release date: | 2018-08-01 | | Last modified: | 2024-03-27 | | Method: | X-RAY DIFFRACTION (2.91 Å) | | Cite: | Enzymatic one-step ring contraction for quinolone biosynthesis.
Nat Commun, 9, 2018
|
|
5YY3
 
 | |
7CP6
 
 | | Crystal structure of FqzB | | Descriptor: | FLAVIN-ADENINE DINUCLEOTIDE, IODIDE ION, MAK1-like monooxygenase, ... | | Authors: | Hara, K, Hashimoto, H, Matsushita, T, Kishimoto, S, Watanabe, K. | | Deposit date: | 2020-08-06 | | Release date: | 2020-12-30 | | Last modified: | 2021-01-13 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Structural and Functional Analyses of a Spiro-Carbon-Forming, Highly Promiscuous Epoxidase from Fungal Natural Product Biosynthesis.
Biochemistry, 59, 2020
|
|
7CP7
 
 | | Crystal structure of FqzB, native proteins | | Descriptor: | FLAVIN-ADENINE DINUCLEOTIDE, IODIDE ION, MAK1-like monooxygenase | | Authors: | Hara, K, Hashimoto, H, Matsushita, T, Kishimoto, S, Watanabe, K. | | Deposit date: | 2020-08-06 | | Release date: | 2020-12-30 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Structural and Functional Analyses of a Spiro-Carbon-Forming, Highly Promiscuous Epoxidase from Fungal Natural Product Biosynthesis.
Biochemistry, 59, 2020
|
|
