7N1X
 
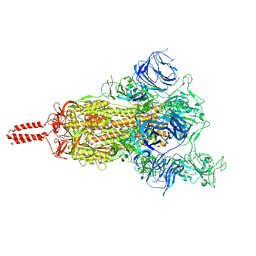 | | Structural basis for enhanced infectivity and immune evasion of SARS-CoV-2 variants | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-[alpha-L-fucopyranose-(1-6)]2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Zhang, J, Cai, Y.F, Xiao, T.S, Rawson, S, Peng, H.Q, Sterling, S.M, Walsh Jr, R.M, Volloch, S.R, Chen, B. | | Deposit date: | 2021-05-28 | | Release date: | 2021-07-07 | | Last modified: | 2024-10-30 | | Method: | ELECTRON MICROSCOPY (4 Å) | | Cite: | Structural basis for enhanced infectivity and immune evasion of SARS-CoV-2 variants.
Science, 373, 2021
|
|
7N1T
 
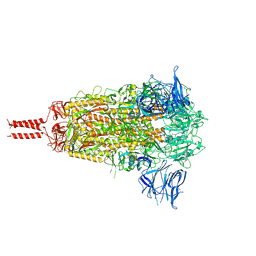 | | Structural basis for enhanced infectivity and immune evasion of SARS-CoV-2 variants | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-[alpha-L-fucopyranose-(1-6)]2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Zhang, J, Cai, Y.F, Xiao, T.S, Rawson, S, Peng, H.Q, Sterling, S.M, Walsh Jr, R.M, Volloch, S.R, Chen, B. | | Deposit date: | 2021-05-28 | | Release date: | 2021-07-07 | | Last modified: | 2021-08-18 | | Method: | ELECTRON MICROSCOPY (3.44 Å) | | Cite: | Structural basis for enhanced infectivity and immune evasion of SARS-CoV-2 variants.
Science, 373, 2021
|
|
7N1Q
 
 | | Structural basis for enhanced infectivity and immune evasion of SARS-CoV-2 variants | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-[alpha-L-fucopyranose-(1-6)]2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Zhang, J, Cai, Y.F, Xiao, T.S, Rawson, S, Peng, H.Q, Sterling, S.M, Walsh Jr, R.M, Volloch, S.R, Chen, B. | | Deposit date: | 2021-05-28 | | Release date: | 2021-07-07 | | Last modified: | 2021-08-18 | | Method: | ELECTRON MICROSCOPY (3.44 Å) | | Cite: | Structural basis for enhanced infectivity and immune evasion of SARS-CoV-2 variants.
Science, 373, 2021
|
|
7N1V
 
 | | Structural basis for enhanced infectivity and immune evasion of SARS-CoV-2 variants | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-[alpha-L-fucopyranose-(1-6)]2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Zhang, J, Cai, Y.F, Xiao, T.S, Rawson, S, Peng, H.Q, Sterling, S.M, Walsh Jr, R.M, Volloch, S.R, Chen, B. | | Deposit date: | 2021-05-28 | | Release date: | 2021-07-07 | | Last modified: | 2024-10-30 | | Method: | ELECTRON MICROSCOPY (3.21 Å) | | Cite: | Structural basis for enhanced infectivity and immune evasion of SARS-CoV-2 variants.
Science, 373, 2021
|
|
6XRA
 
 | | Distinct conformational states of SARS-CoV-2 spike protein | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Spike glycoprotein, ... | | Authors: | Zhang, J, Cai, Y.F, Xiao, T.S, Peng, H.Q, Sterling, S.M, Walsh Jr, R.M, Rawson, S, Rits-Volloch, S, Chen, B. | | Deposit date: | 2020-07-11 | | Release date: | 2020-07-22 | | Last modified: | 2024-10-30 | | Method: | ELECTRON MICROSCOPY (3 Å) | | Cite: | Distinct conformational states of SARS-CoV-2 spike protein.
Science, 369, 2020
|
|
6XR8
 
 | | Distinct conformational states of SARS-CoV-2 spike protein | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-[alpha-L-fucopyranose-(1-6)]2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Zhang, J, Cai, Y.F, Xiao, T.S, Peng, H.Q, Sterling, S.M, Walsh Jr, R.M, Rawson, S, Volloch, S.R, Chen, B. | | Deposit date: | 2020-07-11 | | Release date: | 2020-07-22 | | Last modified: | 2020-11-25 | | Method: | ELECTRON MICROSCOPY (2.9 Å) | | Cite: | Distinct conformational states of SARS-CoV-2 spike protein.
Science, 369, 2020
|
|
6Y5U
 
 | |
6Y56
 
 | |
7E17
 
 | | Structure of dimeric uPAR | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, Urokinase plasminogen activator surface receptor | | Authors: | Cai, Y, Huang, M. | | Deposit date: | 2021-02-01 | | Release date: | 2021-12-22 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.96 Å) | | Cite: | Crystal structure and cellular functions of uPAR dimer
Nat Commun, 13, 2022
|
|
8AZT
 
 | | Type II amyloid-beta 42 filaments from high-spin supernatants of aqueous extracts from Alzheimer's disease brains | ABeta42 | | Descriptor: | Amyloid-beta precursor protein | | Authors: | Yang, Y, Stern, M.A, Meunier, L.A, Liu, W, Cai, Y.Q, Ericsson, M, Liu, L, Selkoe, J.D, Goedert, M, Scheres, H.W.S. | | Deposit date: | 2022-09-06 | | Release date: | 2022-11-02 | | Last modified: | 2024-07-24 | | Method: | ELECTRON MICROSCOPY (3.7 Å) | | Cite: | Abundant A beta fibrils in ultracentrifugal supernatants of aqueous extracts from Alzheimer's disease brains.
Neuron, 111, 2023
|
|
8AZU
 
 | | Paired helical tau filaments from high-spin supernatants of aqueous extracts from Alzheimer's disease brains | PHF Tau | | Descriptor: | Microtubule-associated protein tau | | Authors: | Yang, Y, Stern, M.A, Meunier, L.A, Liu, W, Cai, Y.Q, Ericsson, M, Liu, L, Selkoe, J.D, Goedert, M, Scheres, H.W.S. | | Deposit date: | 2022-09-06 | | Release date: | 2023-05-24 | | Last modified: | 2024-07-24 | | Method: | ELECTRON MICROSCOPY (3.1 Å) | | Cite: | Abundant A beta fibrils in ultracentrifugal supernatants of aqueous extracts from Alzheimer's disease brains.
Neuron, 111, 2023
|
|
5DP4
 
 | | Crystal Structure of EV71 3C Proteinase in complex with compound 3 | | Descriptor: | 3C proteinase, ethyl (2Z,4S)-4-{[(2S)-2-methyl-3-phenylpropanoyl]amino}-5-[(3S)-2-oxopyrrolidin-3-yl]pent-2-enoate | | Authors: | Wu, C, Zhang, L, Li, P, Cai, Q, Peng, X, Li, N, Cai, Y, Li, J, Lin, T. | | Deposit date: | 2015-09-12 | | Release date: | 2016-03-30 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.21 Å) | | Cite: | Fragment-wise design of inhibitors to 3C proteinase from enterovirus 71
Biochim.Biophys.Acta, 1860, 2016
|
|
5DP9
 
 | | Crystal Structure of EV71 3C Proteinase in complex with compound 9 | | Descriptor: | 3C proteinase, ethyl (2Z,4S)-4-[(N-{[(cyclobutylmethyl)amino](oxo)acetyl}-L-phenylalanyl)amino]-5-[(3S)-2-oxopyrrolidin-3-yl]pent-2-enoate | | Authors: | Wu, C, Zhang, L, Li, P, Cai, Q, Peng, X, Li, N, Cai, Y, Li, J, Lin, T. | | Deposit date: | 2015-09-12 | | Release date: | 2016-03-30 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Fragment-wise design of inhibitors to 3C proteinase from enterovirus 71
Biochim.Biophys.Acta, 1860, 2016
|
|
5DP8
 
 | | Crystal Structure of EV71 3C Proteinase in complex with compound 8 | | Descriptor: | 3C proteinase, ethyl (2Z,4S)-4-[(N-{[(2-cyclopropylethyl)amino](oxo)acetyl}-L-phenylalanyl)amino]-5-[(3S)-2-oxopyrrolidin-3-yl]pent-2-enoate | | Authors: | Wu, C, Zhang, L, Li, P, Cai, Q, Peng, X, Li, N, Cai, Y, Li, J, Lin, T. | | Deposit date: | 2015-09-12 | | Release date: | 2016-03-30 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Fragment-wise design of inhibitors to 3C proteinase from enterovirus 71
Biochim.Biophys.Acta, 1860, 2016
|
|
5DPA
 
 | | Crystal Structure of EV71 3C Proteinase in complex with compound 6 | | Descriptor: | 3C proteinase, ethyl (2Z,4S)-4-[(N-acetyl-L-phenylalanyl)amino]-5-[(3S)-2-oxopyrrolidin-3-yl]pent-2-enoate | | Authors: | Wu, C, Zhang, L, Li, P, Cai, Q, Peng, X, Li, N, Cai, Y, Li, J, Lin, T. | | Deposit date: | 2015-09-12 | | Release date: | 2016-03-30 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.06 Å) | | Cite: | Fragment-wise design of inhibitors to 3C proteinase from enterovirus 71
Biochim.Biophys.Acta, 1860, 2016
|
|
5DP6
 
 | | Crystal Structure of EV71 3C Proteinase in complex with compound 7 | | Descriptor: | 3C proteinase, ethyl (2Z,4S)-4-{[N-(3-cyclopropylpropanoyl)-L-phenylalanyl]amino}-5-[(3S)-2-oxopyrrolidin-3-yl]pent-2-enoate | | Authors: | Wu, C, Zhang, L, Li, P, Cai, Q, Peng, X, Li, N, Cai, Y, Li, J, Lin, T. | | Deposit date: | 2015-09-12 | | Release date: | 2016-03-30 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (3.01 Å) | | Cite: | Fragment-wise design of inhibitors to 3C proteinase from enterovirus 71
Biochim.Biophys.Acta, 1860, 2016
|
|
5DP3
 
 | | Crystal Structure of EV71 3C Proteinase in complex with compound 2 | | Descriptor: | 3C proteinase, ethyl (4S)-5-[(3S)-2-oxopyrrolidin-3-yl]-4-[(3-phenylpropanoyl)amino]pent-2-enoate | | Authors: | Wu, C, Zhang, L, Li, P, Cai, Q, Peng, X, Li, N, Cai, Y, Li, J, Lin, T. | | Deposit date: | 2015-09-12 | | Release date: | 2016-03-30 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (2.05 Å) | | Cite: | Fragment-wise design of inhibitors to 3C proteinase from enterovirus 71
Biochim.Biophys.Acta, 1860, 2016
|
|
5DP5
 
 | | Crystal Structure of EV71 3C Proteinase in complex with compound 4 | | Descriptor: | 3C proteinase, ethyl (2Z,4S)-4-{[(2R,5S)-5-amino-2-(4-fluorobenzyl)-6-methyl-4-oxoheptanoyl]amino}-5-[(3S)-2-oxopyrrolidin-3-yl]pent-2-enoate | | Authors: | Wu, C, Zhang, L, Li, P, Cai, Q, Peng, X, Li, N, Cai, Y, Li, J, Lin, T. | | Deposit date: | 2015-09-12 | | Release date: | 2016-03-30 | | Last modified: | 2016-04-06 | | Method: | X-RAY DIFFRACTION (2.03 Å) | | Cite: | Fragment-wise design of inhibitors to 3C proteinase from enterovirus 71
Biochim.Biophys.Acta, 1860, 2016
|
|
5DP7
 
 | | Crystal Structure of EV71 3C Proteinase in complex with compound 5 | | Descriptor: | 3C proteinase, ethyl (2Z,4S)-4-{[N-(3-methylbutanoyl)-L-phenylalanyl]amino}-5-[(3S)-2-oxopyrrolidin-3-yl]pent-2-enoate | | Authors: | Wu, C, Zhang, L, Li, P, Cai, Q, Peng, X, Li, N, Cai, Y, Li, J, Lin, T. | | Deposit date: | 2015-09-12 | | Release date: | 2016-03-30 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (2.08 Å) | | Cite: | Fragment-wise design of inhibitors to 3C proteinase from enterovirus 71
Biochim.Biophys.Acta, 1860, 2016
|
|
7M4R
 
 | |
7BQK
 
 | | The structure of PdxI in complex with its substrate analogue | | Descriptor: | 1,2-ETHANEDIOL, 3-[(E,2S,4S)-2,4-dimethyloct-6-enoyl]-4-oxidanyl-1H-pyridin-2-one, GLYCEROL, ... | | Authors: | Cai, Y.J, Ohashi, M, Zhou, J.H, Tang, Y. | | Deposit date: | 2020-03-24 | | Release date: | 2020-10-14 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (1.99 Å) | | Cite: | An enzymatic Alder-ene reaction.
Nature, 586, 2020
|
|
7BQL
 
 | | The crystal structure of PdxI complex with the Alder-ene adduct | | Descriptor: | 3-[(1R,2S,4R,6S)-2-ethenyl-4,6-dimethyl-cyclohexyl]-4-oxidanyl-1H-pyridin-2-one, DI(HYDROXYETHYL)ETHER, Methyltransf_2 domain-containing protein | | Authors: | Cai, Y.J, Ohashi, M, Zhou, J.H, Tang, Y. | | Deposit date: | 2020-03-24 | | Release date: | 2020-10-14 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.396 Å) | | Cite: | An enzymatic Alder-ene reaction.
Nature, 586, 2020
|
|
7BQJ
 
 | | The structure of PdxI | | Descriptor: | 1,2-ETHANEDIOL, GLYCEROL, Methyltransf_2 domain-containing protein, ... | | Authors: | Cai, Y.J, Ohashi, M, Zhou, J.H, Tang, Y. | | Deposit date: | 2020-03-24 | | Release date: | 2020-10-14 | | Last modified: | 2024-03-27 | | Method: | X-RAY DIFFRACTION (1.98 Å) | | Cite: | An enzymatic Alder-ene reaction.
Nature, 586, 2020
|
|
7BQO
 
 | | The structure of HpiI in complex with its substrate analogue | | Descriptor: | 2-[3-(2-HYDROXY-1,1-DIHYDROXYMETHYL-ETHYLAMINO)-PROPYLAMINO]-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, 3-[(E,2S,4S)-2,4-dimethyloct-6-enoyl]-4-oxidanyl-1H-pyridin-2-one, GLYCEROL, ... | | Authors: | Cai, Y.J, Ohashi, M, Zhou, J.H, Tang, Y. | | Deposit date: | 2020-03-25 | | Release date: | 2020-10-14 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (1.53 Å) | | Cite: | An enzymatic Alder-ene reaction.
Nature, 586, 2020
|
|
7BQP
 
 | | The structure of HpiI | | Descriptor: | (4S)-2-METHYL-2,4-PENTANEDIOL, 1,2-ETHANEDIOL, GLYCEROL, ... | | Authors: | Cai, Y.J, Ohashi, M, Zhou, J.H, Tang, Y. | | Deposit date: | 2020-03-25 | | Release date: | 2020-10-14 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (1.33 Å) | | Cite: | An enzymatic Alder-ene reaction.
Nature, 586, 2020
|
|
