1DLQ
 
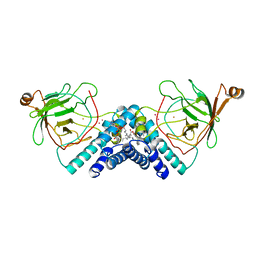 | | STRUCTURE OF CATECHOL 1,2-DIOXYGENASE FROM ACINETOBACTER SP. ADP1 INHIBITED BY BOUND MERCURY | | Descriptor: | CATECHOL 1,2-DIOXYGENASE, FE (III) ION, MERCURY (II) ION, ... | | Authors: | Vetting, M.W, Ohlendorf, D.H. | | Deposit date: | 1999-12-11 | | Release date: | 2000-05-23 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | The 1.8 A crystal structure of catechol 1,2-dioxygenase reveals a novel hydrophobic helical zipper as a subunit linker.
Structure Fold.Des., 8, 2000
|
|
1MHZ
 
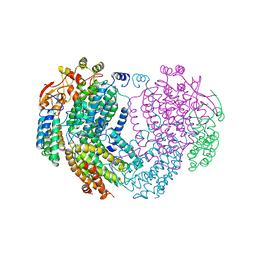 | | METHANE MONOOXYGENASE HYDROXYLASE | | Descriptor: | FE (III) ION, METHANE MONOOXYGENASE HYDROXYLASE | | Authors: | Elango, N, Radhakrishnan, R, Froland, W.A, Waller, B.J, Earhart, C.A, Lipscomb, J.D, Ohlendorf, D.H. | | Deposit date: | 1996-10-21 | | Release date: | 1997-05-15 | | Last modified: | 2024-05-22 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Crystal structure of the hydroxylase component of methane monooxygenase from Methylosinus trichosporium OB3b
Protein Sci., 6, 1997
|
|
1YKK
 
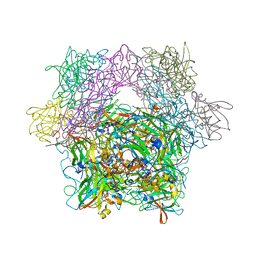 | | Protocatechuate 3,4-Dioxygenase Y408C Mutant | | Descriptor: | FE (III) ION, Protocatechuate 3,4-dioxygenase alpha chain, Protocatechuate 3,4-dioxygenase beta chain | | Authors: | Brown, C.K, Ohlendorf, D.H. | | Deposit date: | 2005-01-18 | | Release date: | 2005-08-16 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (2.06 Å) | | Cite: | Roles of the equatorial tyrosyl iron ligand of protocatechuate 3,4-dioxygenase in catalysis
Biochemistry, 44, 2005
|
|
1Q0O
 
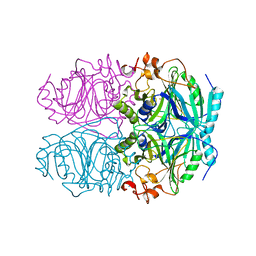 | | CRYSTAL STRUCTURE OF HOMOPROTOCATECHUATE 2,3-DIOXYGENASE FROM BREVIBACTERIUM FUSCUM (FULL LENGTH PROTEIN) | | Descriptor: | FE (III) ION, homoprotocatechuate 2,3-dioxygenase | | Authors: | Vetting, M.W, Wackett, L.P, Que, L, Lipscomb, J.D, Ohlendorf, D.H. | | Deposit date: | 2003-07-16 | | Release date: | 2003-07-29 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Crystallographic comparison of manganese- and iron-dependent homoprotocatechuate 2,3-dioxygenases.
J.Bacteriol., 186, 2004
|
|
1QTF
 
 | | CRYSTAL STRUCTURE OF EXFOLIATIVE TOXIN B | | Descriptor: | EXFOLIATIVE TOXIN B | | Authors: | Vath, G.M, Earhart, C.A, Monie, D.D, Schlievert, P.M, Ohlendorf, D.H. | | Deposit date: | 1999-06-27 | | Release date: | 1999-08-31 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | The crystal structure of exfoliative toxin B: a superantigen with enzymatic activity.
Biochemistry, 38, 1999
|
|
1Q0C
 
 | | Anerobic Substrate Complex of Homoprotocatechuate 2,3-Dioxygenase from Brevibacterium fuscum. (Complex with 3,4-Dihydroxyphenylacetate) | | Descriptor: | 2-(3,4-DIHYDROXYPHENYL)ACETIC ACID, FE (III) ION, homoprotocatechuate 2,3-dioxygenase | | Authors: | Vetting, M.W, Wackett, L.P, Que, L, Lipscomb, J.D, Ohlendorf, D.H. | | Deposit date: | 2003-07-15 | | Release date: | 2003-07-29 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Crystallographic comparison of manganese- and iron-dependent homoprotocatechuate 2,3-dioxygenases.
J.Bacteriol., 186, 2004
|
|
3HK1
 
 | | Identification and Characterization of a Small Molecule Inhibitor of Fatty Acid Binding Proteins | | Descriptor: | 4-{[2-(methoxycarbonyl)-5-(2-thienyl)-3-thienyl]amino}-4-oxo-2-butenoic acid, Fatty acid-binding protein, adipocyte | | Authors: | Hertzel, A.V, Hellberg, K, Reynolds, J.M, Kruse, A.C, Juhlmann, B.E, Smith, A.J, Sanders, M.A, Ohlendorf, D.H, Suttles, J, Bernlohr, D.A. | | Deposit date: | 2009-05-22 | | Release date: | 2009-09-22 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Identification and characterization of a small molecule inhibitor of Fatty Acid binding proteins.
J.Med.Chem., 52, 2009
|
|
3HL6
 
 | | Staphylococcus aureus pathogenicity island 3 ORF9 protein | | Descriptor: | CHLORIDE ION, Pathogenicity island protein | | Authors: | Kruse, A.C, Huseby, M, Shi, K, Digre, J, Schlievert, P.M, Ohlendorf, D.H, Earhart, C.A. | | Deposit date: | 2009-05-26 | | Release date: | 2010-06-09 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Structural and functional studies of a pathogenicity island protein from Staphylococcus aureus
To be Published
|
|
3JS1
 
 | | Crystal structure of adipocyte fatty acid binding protein covalently modified with 4-hydroxy-2-nonenal | | Descriptor: | Adipocyte fatty acid-binding protein, PHOSPHATE ION | | Authors: | Hellberg, K, Grimsrud, P.A, Kruse, A.C, Banaszak, L.J, Ohlendorf, D.H, Bernlohr, D.A. | | Deposit date: | 2009-09-09 | | Release date: | 2010-08-25 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (1.81 Å) | | Cite: | X-ray crystallographic analysis of adipocyte fatty acid binding protein (aP2) modified with 4-hydroxy-2-nonenal.
Protein Sci., 19, 2010
|
|
3JSQ
 
 | | Crystal structure of adipocyte fatty acid binding protein non-covalently modified with 4-hydroxy-2-nonenal | | Descriptor: | (2E,4R)-4-HYDROXYNON-2-ENAL, Adipocyte fatty acid-binding protein, CHLORIDE ION, ... | | Authors: | Hellberg, K, Grimsrud, P.A, Kruse, A.C, Banaszak, L.J, Ohlendorf, D.H, Bernlohr, D.A. | | Deposit date: | 2009-09-10 | | Release date: | 2010-08-25 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | X-ray crystallographic analysis of adipocyte fatty acid binding protein (aP2) modified with 4-hydroxy-2-nonenal.
Protein Sci., 19, 2010
|
|
3K55
 
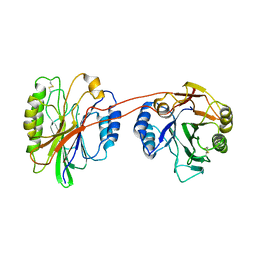 | | Structure of beta hairpin deletion mutant of beta toxin from Staphylococcus aureus | | Descriptor: | Beta-hemolysin, CHLORIDE ION, SODIUM ION | | Authors: | Kruse, A.C, Huseby, M, Shi, K, Digre, J, Ohlendorf, D.H, Earhart, C.A. | | Deposit date: | 2009-10-06 | | Release date: | 2011-01-26 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (3.35 Å) | | Cite: | Structure of a mutant beta toxin from Staphylococcus aureus reveals domain swapping and conformational flexibility
Acta Crystallogr.,Sect.F, 67, 2011
|
|
2AZQ
 
 | | Crystal Structure of Catechol 1,2-Dioxygenase from Pseudomonas arvilla C-1 | | Descriptor: | 1,2-DIACYL-SN-GLYCERO-3-PHOSHOCHOLINE, FE (III) ION, catechol 1,2-dioxygenase | | Authors: | Earhart, C.A, Vetting, M.W, Gosu, R, Michaud-Soret, I, Que, L, Ohlendorf, D.H. | | Deposit date: | 2005-09-12 | | Release date: | 2005-10-04 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (2.65 Å) | | Cite: | Structure of catechol 1,2-dioxygenase from Pseudomonas arvilla
Biochem.Biophys.Res.Commun., 338, 2005
|
|
1YKL
 
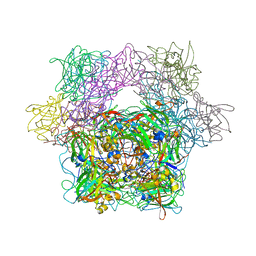 | | Protocatechuate 3,4-Dioxygenase Y408C mutant bound to DHB | | Descriptor: | 3,4-DIHYDROXYBENZOIC ACID, FE (III) ION, Protocatechuate 3,4-dioxygenase alpha chain, ... | | Authors: | Brown, C.K, Ohlendorf, D.H. | | Deposit date: | 2005-01-18 | | Release date: | 2005-08-16 | | Last modified: | 2021-10-20 | | Method: | X-RAY DIFFRACTION (2.25 Å) | | Cite: | Roles of the equatorial tyrosyl iron ligand of protocatechuate 3,4-dioxygenase in catalysis
Biochemistry, 44, 2005
|
|
1YKM
 
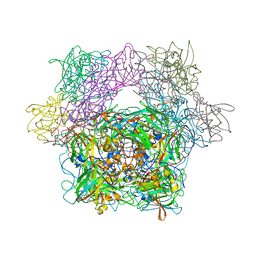 | | Protocatechuate 3,4-Dioxygenase Y408E mutant | | Descriptor: | FE (III) ION, Protocatechuate 3,4-dioxygenase alpha chain, Protocatechuate 3,4-dioxygenase beta chain | | Authors: | Brown, C.K, Ohlendorf, D.H. | | Deposit date: | 2005-01-18 | | Release date: | 2005-08-16 | | Last modified: | 2021-10-20 | | Method: | X-RAY DIFFRACTION (2.22 Å) | | Cite: | Roles of the equatorial tyrosyl iron ligand of protocatechuate 3,4-dioxygenase in catalysis
Biochemistry, 44, 2005
|
|
1YKN
 
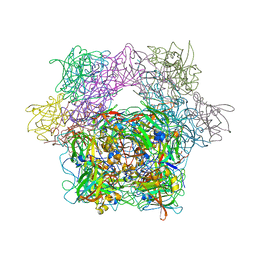 | | Protocatechuate 3,4-dioxygenase Y408E mutant bound to DHB | | Descriptor: | 3,4-DIHYDROXYBENZOIC ACID, FE (III) ION, Protocatechuate 3,4-dioxygenase alpha chain, ... | | Authors: | Brown, C.K, Ohlendorf, D.H. | | Deposit date: | 2005-01-18 | | Release date: | 2005-08-16 | | Last modified: | 2021-10-20 | | Method: | X-RAY DIFFRACTION (2.06 Å) | | Cite: | Roles of the equatorial tyrosyl iron ligand of protocatechuate 3,4-dioxygenase in catalysis
Biochemistry, 44, 2005
|
|
1YKO
 
 | | Protocatechuate 3,4-Dioxygenase Y408H mutant | | Descriptor: | FE (III) ION, Protocatechuate 3,4-dioxygenase alpha chain, Protocatechuate 3,4-dioxygenase beta chain | | Authors: | Brown, C.K, Ohlendorf, D.H. | | Deposit date: | 2005-01-18 | | Release date: | 2005-08-16 | | Last modified: | 2021-10-20 | | Method: | X-RAY DIFFRACTION (2.54 Å) | | Cite: | Roles of the equatorial tyrosyl iron ligand of protocatechuate 3,4-dioxygenase in catalysis
Biochemistry, 44, 2005
|
|
1YKP
 
 | | Protocatechuate 3,4-Dioxygenase Y408H mutant bound to DHB | | Descriptor: | 3,4-DIHYDROXYBENZOIC ACID, FE (III) ION, Protocatechuate 3,4-dioxygenase alpha chain, ... | | Authors: | Brown, C.K, Ohlendorf, D.H. | | Deposit date: | 2005-01-18 | | Release date: | 2005-08-16 | | Last modified: | 2021-10-20 | | Method: | X-RAY DIFFRACTION (2.41 Å) | | Cite: | Roles of the equatorial tyrosyl iron ligand of protocatechuate 3,4-dioxygenase in catalysis
Biochemistry, 44, 2005
|
|
1XF1
 
 | | Structure of C5a peptidase- a key virulence factor from Streptococcus | | Descriptor: | ACETATE ION, C5a peptidase, CALCIUM ION, ... | | Authors: | Brown, C.K, Gu, Z.Y, Cleary, P.P, Matsuka, Y, Olmstead, S, Ohlendorf, D.H, Earhart, C.A. | | Deposit date: | 2004-09-13 | | Release date: | 2005-11-22 | | Last modified: | 2021-10-20 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Structure of the streptococcal cell wall C5a peptidase
Proc.Natl.Acad.Sci.Usa, 102, 2005
|
|
2BUV
 
 | | Crystal Structure Of Protocatechuate 3,4-Dioxygenase from Acinetobacter Sp. ADP1 Mutant R457S in Complex with Protocatechuate | | Descriptor: | 3,4-DIHYDROXYBENZOIC ACID, FE (III) ION, PROTOCATECHUATE 3,4-DIOXYGENASE ALPHA CHAIN, ... | | Authors: | Vetting, M.W, Valley, M.P, D'Argenio, D.A, Ornston, L.N, Lipscomb, J.D, Ohlendorf, D.H. | | Deposit date: | 2005-06-17 | | Release date: | 2006-09-04 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Biophysical Analyses of Designed and Selected Mutants of Protocatechuate 3,4-Dioxygenase
Annu.Rev.Microbiol., 58, 2004
|
|
2BUR
 
 | | Crystal Structure Of Wild-Type Protocatechuate 3,4-Dioxygenase from Acinetobacter Sp. ADP1 in Complex with 4-hydroxybenzoate | | Descriptor: | FE (III) ION, P-HYDROXYBENZOIC ACID, PROTOCATECHUATE 3,4-DIOXYGENASE ALPHA CHAIN, ... | | Authors: | Vetting, M.W, Valley, M.P, D'Argenio, D.A, Ornston, L.N, Lipscomb, J.D, Ohlendorf, D.H. | | Deposit date: | 2005-06-17 | | Release date: | 2006-09-05 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Biophysical Analyses of Designed and Selected Mutants of Protocatechuate 3,4-Dioxygenase
Annu.Rev.Microbiol., 58, 2004
|
|
2BUQ
 
 | | Crystal Structure Of Wild-Type Protocatechuate 3,4-Dioxygenase from Acinetobacter Sp. ADP1 in Complex with Catechol | | Descriptor: | CATECHOL, FE (III) ION, PROTOCATECHUATE 3,4-DIOXYGENASE ALPHA CHAIN, ... | | Authors: | Vetting, M.W, Valley, M.P, D'Argenio, D.A, Ornston, L.N, Lipscomb, J.D, Ohlendorf, D.H. | | Deposit date: | 2005-06-17 | | Release date: | 2006-09-05 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Biophysical Analyses of Designed and Selected Mutants of Protocatechuate 3,4-Dioxygenase
Annu.Rev.Microbiol., 58, 2004
|
|
2BUT
 
 | | Crystal Structure Of Protocatechuate 3,4-Dioxygenase from Acinetobacter Sp. ADP1 Mutant R457S - APO | | Descriptor: | FE (III) ION, HYDROXIDE ION, PROTOCATECHUATE 3,4-DIOXYGENASE ALPHA CHAIN, ... | | Authors: | Vetting, M.W, Valley, M.P, D'Argenio, D.A, Ornston, L.N, Lipscomb, J.D, Ohlendorf, D.H. | | Deposit date: | 2005-06-17 | | Release date: | 2006-09-05 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | Biophysical Analyses of Designed and Selected Mutants of Protocatechuate 3,4-Dioxygenase
Annu.Rev.Microbiol., 58, 2004
|
|
2BUM
 
 | | Crystal Structure Of Wild-Type Protocatechuate 3,4-Dioxygenase from Acinetobacter Sp. ADP1 | | Descriptor: | FE (III) ION, HYDROXIDE ION, PROTOCATECHUATE 3,4-DIOXYGENASE ALPHA CHAIN, ... | | Authors: | Vetting, M.W, Valley, M.P, D'Argenio, D.A, Ornston, L.N, Lipscomb, J.D, Ohlendorf, D.H. | | Deposit date: | 2005-06-14 | | Release date: | 2006-09-05 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Biophysical Analyses of Designed and Selected Mutants of Protocatechuate 3,4-Dioxygenase
Annu.Rev.Microbiol., 58, 2004
|
|
2AXU
 
 | | Structure of PrgX | | Descriptor: | PrgX | | Authors: | Shi, K, Brown, C.K, Gu, Z.Y, Kozlowicz, B.K, Dunny, G.M, Ohlendorf, D.H, Earhart, C.A. | | Deposit date: | 2005-09-06 | | Release date: | 2005-12-06 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (2.9 Å) | | Cite: | Structure of peptide sex pheromone receptor PrgX and PrgX/pheromone complexes and regulation of conjugation in Enterococcus faecalis.
Proc.Natl.Acad.Sci.Usa, 102, 2005
|
|
2AWI
 
 | | Structure of PrgX Y153C mutant | | Descriptor: | PrgX | | Authors: | Shi, K, Brown, C.K, Gu, Z.Y, Kozlowicz, B.k, Dunny, G.M, Ohlendorf, D.H, Earhart, C.A. | | Deposit date: | 2005-09-01 | | Release date: | 2005-12-06 | | Last modified: | 2021-10-20 | | Method: | X-RAY DIFFRACTION (2.25 Å) | | Cite: | Structure of peptide sex pheromone receptor PrgX and PrgX/pheromone complexes and regulation of conjugation in Enterococcus faecalis.
Proc.Natl.Acad.Sci.Usa, 102, 2005
|
|
