8OXE
 
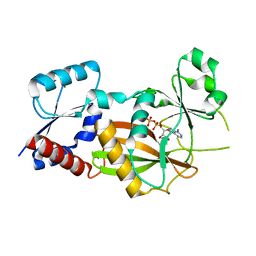 | | Inositol 1,3,4-trisphosphate 5/6-kinase 1 from Solanum tuberosum (StITPK1) in complex with ADP/Mg2+ | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, Inositol-tetrakisphosphate 1-kinase, MAGNESIUM ION | | Authors: | Faba-Rodriguez, R, Li, A.W.H, Brearley, C.A, Hemmings, A.M. | | Deposit date: | 2023-05-02 | | Release date: | 2024-03-27 | | Method: | X-RAY DIFFRACTION (2.26 Å) | | Cite: | Crystal Structure and Enzymology of Solanum tuberosum Inositol Tris/Tetrakisphosphate Kinase 1 ( St ITPK1).
Biochemistry, 63, 2024
|
|
1UCO
 
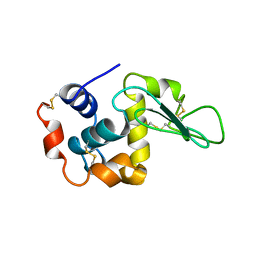 | | HEN EGG-WHITE LYSOZYME, LOW HUMIDITY FORM | | Descriptor: | LYSOZYME | | Authors: | Nagendra, H.G, Sudarsanakumar, C, Vijayan, M. | | Deposit date: | 1995-12-31 | | Release date: | 1996-07-11 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | An X-ray analysis of native monoclinic lysozyme. A case study on the reliability of refined protein structures and a comparison with the low-humidity form in relation to mobility and enzyme action.
Acta Crystallogr.,Sect.D, 52, 1996
|
|
1MNH
 
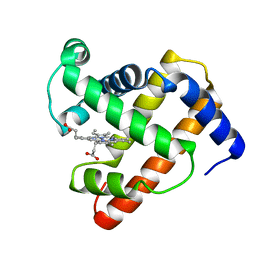 | | INTERACTIONS AMONG RESIDUES CD3, E7, E10 AND E11 IN MYOGLOBINS: ATTEMPTS TO SIMULATE THE O2 AND CO BINDING PROPERTIES OF APLYSIA MYOGLOBIN | | Descriptor: | MYOGLOBIN, PROTOPORPHYRIN IX CONTAINING FE | | Authors: | Davies, G.J, Wilkinson, A.J. | | Deposit date: | 1995-01-11 | | Release date: | 1995-05-08 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Interactions among residues CD3, E7, E10, and E11 in myoglobins: attempts to simulate the ligand-binding properties of Aplysia myoglobin.
Biochemistry, 34, 1995
|
|
1KPT
 
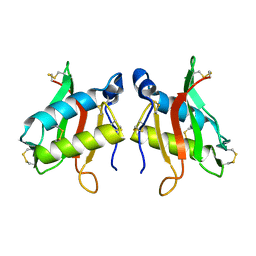 | |
1LWD
 
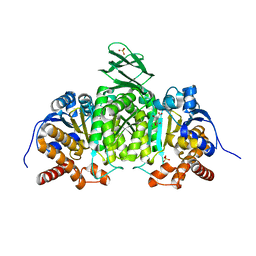 | |
1MNK
 
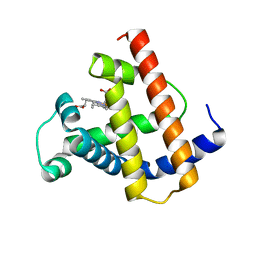 | | INTERACTIONS AMONG RESIDUES CD3, E7, E10 AND E11 IN MYOGLOBINS: ATTEMPTS TO SIMULATE THE O2 AND CO BINDING PROPERTIES OF APLYSIA MYOGLOBIN | | Descriptor: | MYOGLOBIN, PROTOPORPHYRIN IX CONTAINING FE | | Authors: | Krzywda, S, Wilkinson, A.J. | | Deposit date: | 1995-01-11 | | Release date: | 1995-04-20 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Interactions among residues CD3, E7, E10, and E11 in myoglobins: attempts to simulate the ligand-binding properties of Aplysia myoglobin.
Biochemistry, 34, 1995
|
|
1MNJ
 
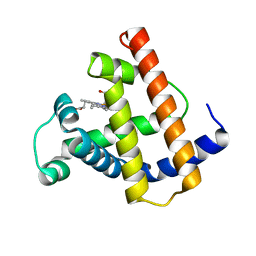 | | INTERACTIONS AMONG RESIDUES CD3, E7, E10 AND E11 IN MYOGLOBINS: ATTEMPTS TO SIMULATE THE O2 AND CO BINDING PROPERTIES OF APLYSIA MYOGLOBIN | | Descriptor: | MYOGLOBIN, PROTOPORPHYRIN IX CONTAINING FE | | Authors: | Krzywda, S, Wilkinson, A.J. | | Deposit date: | 1995-01-11 | | Release date: | 1995-04-20 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Interactions among residues CD3, E7, E10, and E11 in myoglobins: attempts to simulate the ligand-binding properties of Aplysia myoglobin.
Biochemistry, 34, 1995
|
|
1MCY
 
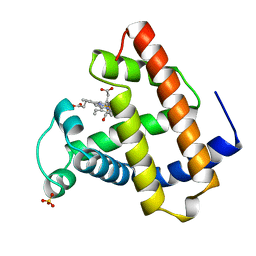 | |
1HN6
 
 | |
1XEK
 
 | |
1XEI
 
 | |
1XEJ
 
 | |
2HLO
 
 | | Crystal Structure of Fragment D-dimer from Human Fibrin Complexed with Gly-hydroxyPro-Arg-Pro-amide | | Descriptor: | 2-acetamido-2-deoxy-alpha-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, CALCIUM ION, ... | | Authors: | Doolittle, R.F, Kollman, J.M, Chen, A, Pandi, L. | | Deposit date: | 2006-07-08 | | Release date: | 2007-07-17 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: |
|
|
2HPC
 
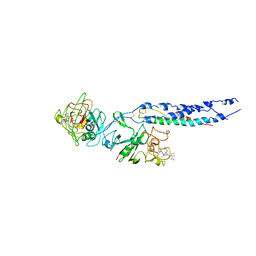 | | Crystal structure of fragment D from Human Fibrinogen Complexed with Gly-Pro-Arg-Pro-amide. | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, CALCIUM ION, Fibrinogen alpha chain, ... | | Authors: | Doolittle, R.F, Kollman, J.M, Chen, A, Pandi, L. | | Deposit date: | 2006-07-17 | | Release date: | 2007-05-29 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (2.9 Å) | | Cite: |
|
|
2HOD
 
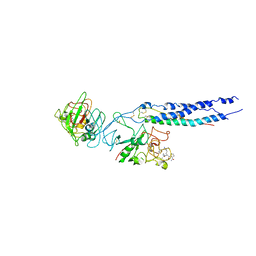 | | Crystal Structure of Fragment D from Human Fibrinogen Complexed with Gly-hydroxyPro-Arg-Pro-amide | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, CALCIUM ION, Fibrinogen alpha chain, ... | | Authors: | Doolittle, R.F, Kollman, J.M, Chen, A, Pandi, L. | | Deposit date: | 2006-07-14 | | Release date: | 2007-05-29 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (2.9 Å) | | Cite: |
|
|
2K9S
 
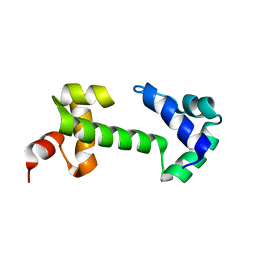 | |
3II9
 
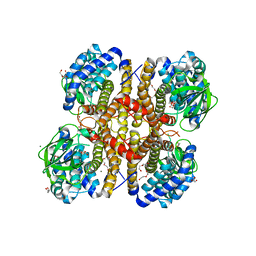 | | Crystal structure of glutaryl-coa dehydrogenase from Burkholderia pseudomallei at 1.73 Angstrom | | Descriptor: | DI(HYDROXYETHYL)ETHER, GLYCEROL, Glutaryl-CoA dehydrogenase, ... | | Authors: | Ismagilov, R.F, Li, L, Du, W.B, Staker, B, Accelerated Technologies Center for Gene to 3D Structure (ATCG3D), Seattle Structural Genomics Center for Infectious Disease (SSGCID) | | Deposit date: | 2009-07-31 | | Release date: | 2009-12-15 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (1.74 Å) | | Cite: | User-loaded SlipChip for equipment-free multiplexed nanoliter-scale experiments.
J.Am.Chem.Soc., 132, 2010
|
|
3IQS
 
 | | Crystal structure of the anti-viral APOBEC3G catalytic domain | | Descriptor: | DNA dC->dU-editing enzyme APOBEC-3G, ZINC ION | | Authors: | Holden, L.G, Prochnow, C, Chang, Y.P, Bransteitter, R, Chelico, L, Sen, U, Stevens, R.C, Goodman, R.F, Chen, X.S. | | Deposit date: | 2009-08-20 | | Release date: | 2009-11-10 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Crystal structure of the anti-viral APOBEC3G catalytic domain and functional implications.
Nature, 456, 2008
|
|
193L
 
 | | THE 1.33 A STRUCTURE OF TETRAGONAL HEN EGG WHITE LYSOZYME | | Descriptor: | CHLORIDE ION, LYSOZYME, SODIUM ION | | Authors: | Vaney, M.C, Maignan, S, Ries-Kautt, M, Ducruix, A. | | Deposit date: | 1995-09-01 | | Release date: | 1995-12-07 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (1.33 Å) | | Cite: | High-resolution structure (1.33 A) of a HEW lysozyme tetragonal crystal grown in the APCF apparatus. Data and structural comparison with a crystal grown under microgravity from SpaceHab-01 mission.
Acta Crystallogr.,Sect.D, 52, 1996
|
|
1BLC
 
 | |
1BMP
 
 | | BONE MORPHOGENETIC PROTEIN-7 | | Descriptor: | BONE MORPHOGENETIC PROTEIN-7 | | Authors: | Griffith, D.L, Scott, D.L. | | Deposit date: | 1995-12-14 | | Release date: | 1997-07-23 | | Last modified: | 2024-11-20 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Three-dimensional structure of recombinant human osteogenic protein 1: structural paradigm for the transforming growth factor beta superfamily.
Proc.Natl.Acad.Sci.USA, 93, 1996
|
|
1D6O
 
 | | NATIVE FKBP | | Descriptor: | AMMONIUM ION, PROTEIN (FK506-BINDING PROTEIN), SULFATE ION | | Authors: | Burkhard, P, Taylor, P, Walkinshaw, M.D. | | Deposit date: | 1999-10-15 | | Release date: | 1999-10-21 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | X-ray structures of small ligand-FKBP complexes provide an estimate for hydrophobic interaction energies.
J.Mol.Biol., 295, 2000
|
|
194L
 
 | | THE 1.40 A STRUCTURE OF SPACEHAB-01 HEN EGG WHITE LYSOZYME | | Descriptor: | CHLORIDE ION, LYSOZYME, SODIUM ION | | Authors: | Vaney, M.C, Maignan, S, Ries-Kautt, M, Ducruix, A. | | Deposit date: | 1995-09-01 | | Release date: | 1995-12-07 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (1.4 Å) | | Cite: | High-resolution structure (1.33 A) of a HEW lysozyme tetragonal crystal grown in the APCF apparatus. Data and structural comparison with a crystal grown under microgravity from SpaceHab-01 mission.
Acta Crystallogr.,Sect.D, 52, 1996
|
|
1D7H
 
 | | FKBP COMPLEXED WITH DMSO | | Descriptor: | AMMONIUM ION, DIMETHYL SULFOXIDE, PROTEIN (FK506-BINDING PROTEIN), ... | | Authors: | Burkhard, P, Taylor, P, Walkinshaw, M.D. | | Deposit date: | 1999-10-18 | | Release date: | 1999-10-21 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | X-ray structures of small ligand-FKBP complexes provide an estimate for hydrophobic interaction energies.
J.Mol.Biol., 295, 2000
|
|
1D7J
 
 | | FKBP COMPLEXED WITH 4-HYDROXY-2-BUTANONE | | Descriptor: | 4-HYDROXY-2-BUTANONE, AMMONIUM ION, PROTEIN (FK506-BINDING PROTEIN), ... | | Authors: | Burkhard, P, Taylor, P, Walkinshaw, M.D. | | Deposit date: | 1999-10-18 | | Release date: | 1999-10-21 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | X-ray structures of small ligand-FKBP complexes provide an estimate for hydrophobic interaction energies.
J.Mol.Biol., 295, 2000
|
|
