1C1E
 
 | | CRYSTAL STRUCTURE OF A DIELS-ALDERASE CATALYTIC ANTIBODY 1E9 IN COMPLEX WITH ITS HAPTEN | | Descriptor: | 1,7,8,9,10,10-HEXACHLORO-4-METHYL-4-AZA-TRICYCLO[5.2.1.0(2,6)]DEC-8-ENE-3,5-DIONE, CATALYTIC ANTIBODY 1E9 (HEAVY CHAIN), CATALYTIC ANTIBODY 1E9 (LIGHT CHAIN), ... | | Authors: | Xu, J, Wilson, I.A. | | Deposit date: | 1999-07-22 | | Release date: | 2000-03-01 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Evolution of shape complementarity and catalytic efficiency from a primordial antibody template.
Science, 286, 1999
|
|
1C16
 
 | | CRYSTAL STRUCTURE ANALYSIS OF THE GAMMA/DELTA T CELL LIGAND T22 | | Descriptor: | MHC-LIKE PROTEIN T22, PROTEIN (BETA-2-MICROGLOBULIN) | | Authors: | Wingren, C, Crowley, M.P, Degano, M, Chien, Y, Wilson, I.A. | | Deposit date: | 1999-07-20 | | Release date: | 2000-01-26 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (3.1 Å) | | Cite: | Crystal structure of a gammadelta T cell receptor ligand T22: a truncated MHC-like fold.
Science, 287, 2000
|
|
1C5B
 
 | | DECARBOXYLASE CATALYTIC ANTIBODY 21D8 UNLIGANDED FORM | | Descriptor: | CHIMERIC DECARBOXYLASE ANTIBODY 21D8 | | Authors: | Hotta, K, Wilson, I.A. | | Deposit date: | 1999-11-08 | | Release date: | 2000-10-11 | | Last modified: | 2024-11-20 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Catalysis of decarboxylation by a preorganized heterogeneous microenvironment: crystal structures of abzyme 21D8.
J.Mol.Biol., 302, 2000
|
|
1C5C
 
 | | DECARBOXYLASE CATALYTIC ANTIBODY 21D8-HAPTEN COMPLEX | | Descriptor: | 2-ACETYLAMINO-NAPTHALENE-1,5-DISULFONIC ACID, CHIMERIC DECARBOXYLASE ANTIBODY 21D8, GLYCEROL | | Authors: | Hotta, K, Wilson, I.A. | | Deposit date: | 1999-11-09 | | Release date: | 2000-10-11 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (1.61 Å) | | Cite: | Catalysis of decarboxylation by a preorganized heterogeneous microenvironment: crystal structures of abzyme 21D8.
J.Mol.Biol., 302, 2000
|
|
3OAZ
 
 | | A non-self sugar mimic of the HIV glycan shield shows enhanced antigenicity | | Descriptor: | CHLORIDE ION, Fab 2G12, heavy chain, ... | | Authors: | Doores, K.J, Fulton, Z, Hong, V, Patel, M.K, Scanlan, C.N, Wormald, M.R, Finn, M.G, Burton, D.R, Wilson, I.A, Davis, B.G. | | Deposit date: | 2010-08-06 | | Release date: | 2011-01-12 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (1.75 Å) | | Cite: | A nonself sugar mimic of the HIV glycan shield shows enhanced antigenicity.
Proc.Natl.Acad.Sci.USA, 107, 2010
|
|
3O2V
 
 | |
3OAY
 
 | | A non-self sugar mimic of the HIV glycan shield shows enhanced antigenicity | | Descriptor: | Fab 2G12, heavy chain, light chain, ... | | Authors: | Doores, K.J, Fulton, Z, Hong, V, Patel, M.K, Scanlan, C.N, Wormald, M.R, Finn, M.G, Burton, D.R, Wilson, I.A, Davis, B.G. | | Deposit date: | 2010-08-05 | | Release date: | 2011-01-12 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | A nonself sugar mimic of the HIV glycan shield shows enhanced antigenicity.
Proc.Natl.Acad.Sci.USA, 107, 2010
|
|
3QG7
 
 | | Structural Basis for Ligand Recognition and Discrimination of a Quorum Quenching Antibody | | Descriptor: | AP4-24H11 Antibody Heavy Chain, AP4-24H11 Antibody Light Chain, HEXAETHYLENE GLYCOL, ... | | Authors: | Kirchdoerfer, R.K, Kaufmann, G.F, Janda, J.D, Wilson, I.A. | | Deposit date: | 2011-01-24 | | Release date: | 2011-03-23 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (2.78 Å) | | Cite: | Structural Basis for Ligand Recognition and Discrimination of a Quorum-quenching Antibody.
J.Biol.Chem., 286, 2011
|
|
2ERJ
 
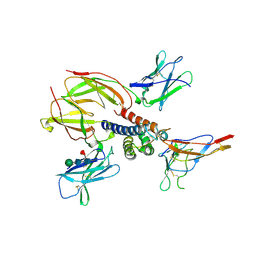 | | Crystal structure of the heterotrimeric interleukin-2 receptor in complex with interleukin-2 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-[alpha-L-fucopyranose-(1-6)]2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Debler, E.W, Stauber, D.J, Wilson, I.A. | | Deposit date: | 2005-10-25 | | Release date: | 2006-02-21 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | Crystal structure of the IL-2 signaling complex: Paradigm for a heterotrimeric cytokine receptor.
Proc.Natl.Acad.Sci.Usa, 103, 2006
|
|
3Q1S
 
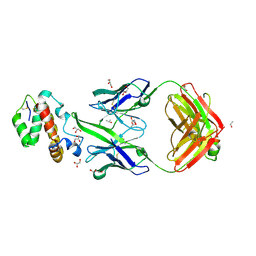 | | HIV-1 neutralizing antibody Z13e1 in complex with epitope display protein | | Descriptor: | GLYCEROL, ISOPROPYL ALCOHOL, Interleukin-22, ... | | Authors: | Stanfield, R.L, Julien, J.-P, Pejchal, R, Gach, J.S, Zwick, M.B, Wilson, I.A. | | Deposit date: | 2010-12-17 | | Release date: | 2011-11-02 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (2.15 Å) | | Cite: | Structure-Based Design of a Protein Immunogen that Displays an HIV-1 gp41 Neutralizing Epitope.
J.Mol.Biol., 414, 2011
|
|
3QQO
 
 | |
3O2W
 
 | |
2FK0
 
 | | Crystal Structure of a H5N1 influenza virus hemagglutinin. | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, beta-D-mannopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, hemagglutinin | | Authors: | Stevens, J, Wilson, I.A. | | Deposit date: | 2006-01-03 | | Release date: | 2006-05-02 | | Last modified: | 2024-11-20 | | Method: | X-RAY DIFFRACTION (2.95 Å) | | Cite: | Structure and Receptor Specificity of the Hemagglutinin from an H5N1 Influenza Virus.
Science, 312, 2006
|
|
3OZ9
 
 | |
3QQI
 
 | |
3QQE
 
 | | Crystal structure of HA2 R106H mutant of H2 hemagglutinin, re-neutralized form | | Descriptor: | 1,2-ETHANEDIOL, 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Xu, R, Wilson, I.A. | | Deposit date: | 2011-02-15 | | Release date: | 2011-03-09 | | Last modified: | 2024-11-27 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Structural Characterization of an Early Fusion Intermediate of Influenza Virus Hemagglutinin.
J.Virol., 85, 2011
|
|
3QQB
 
 | | Crystal structure of HA2 R106H mutant of H2 hemagglutinin, neutral pH form | | Descriptor: | 1,2-ETHANEDIOL, 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Xu, R, Wilson, I.A. | | Deposit date: | 2011-02-15 | | Release date: | 2011-03-09 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (1.97 Å) | | Cite: | Structural Characterization of an Early Fusion Intermediate of Influenza Virus Hemagglutinin.
J.Virol., 85, 2011
|
|
2FL5
 
 | |
3RG1
 
 | | Crystal structure of the RP105/MD-1 complex | | Descriptor: | (1S)-2-{[{[(2R)-2,3-DIHYDROXYPROPYL]OXY}(HYDROXY)PHOSPHORYL]OXY}-1-[(PALMITOYLOXY)METHYL]ETHYL STEARATE, CD180 molecule, LY86 protein, ... | | Authors: | Yoon, S.I, Hong, M, Wilson, I.A. | | Deposit date: | 2011-04-07 | | Release date: | 2011-08-31 | | Last modified: | 2024-11-20 | | Method: | X-RAY DIFFRACTION (2.91 Å) | | Cite: | An unusual dimeric structure and assembly for TLR4 regulator RP105-MD-1.
Nat.Struct.Mol.Biol., 18, 2011
|
|
3R06
 
 | |
3QHF
 
 | |
3QHZ
 
 | | Crystal Structure of human anti-influenza Fab 2D1 | | Descriptor: | Human monoclonal antibody del2D1, Fab Heavy Chain, Fab Light Chain | | Authors: | Ekiert, D.C, Wilson, I.A. | | Deposit date: | 2011-01-26 | | Release date: | 2011-02-16 | | Last modified: | 2024-11-20 | | Method: | X-RAY DIFFRACTION (1.55 Å) | | Cite: | An insertion mutation that distorts antibody binding site architecture enhances function of a human antibody.
MBio, 2, 2011
|
|
3QL0
 
 | | Crystal structure of N23PP/S148A mutant of E. coli dihydrofolate reductase | | Descriptor: | Dihydrofolate reductase, FOLIC ACID, NADP NICOTINAMIDE-ADENINE-DINUCLEOTIDE PHOSPHATE, ... | | Authors: | Bhabha, G, Ekiert, D.C, Wright, P.E, Wilson, I.A. | | Deposit date: | 2011-02-02 | | Release date: | 2011-04-20 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | A dynamic knockout reveals that conformational fluctuations influence the chemical step of enzyme catalysis.
Science, 332, 2011
|
|
2G2R
 
 | |
3QG6
 
 | | Structural Basis for Ligand Recognition and Discrimination of a Quorum Quenching Antibody | | Descriptor: | AP4-24H11 Heavy Chain, AP4-24H11 Light Chain, Agr autoinducing peptide, ... | | Authors: | Kirchdoerfer, R.N, Janda, J.D, Kaufmann, G.F, Wilson, I.A. | | Deposit date: | 2011-01-24 | | Release date: | 2011-03-23 | | Last modified: | 2024-11-27 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Structural Basis for Ligand Recognition and Discrimination of a Quorum-quenching Antibody.
J.Biol.Chem., 286, 2011
|
|
