4ZR7
 
 | | The structure of a domain of a functionally unknown protein from Bacillus subtilis subsp. subtilis str. 168 | | Descriptor: | ACETATE ION, CHLORIDE ION, Sensor histidine kinase ResE | | Authors: | Tan, K, Li, H, Jedrzejczak, R, Joachimiak, A, Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2015-05-11 | | Release date: | 2015-05-27 | | Last modified: | 2024-11-13 | | Method: | X-RAY DIFFRACTION (1.86 Å) | | Cite: | The structure of a domain of a functionally unknown protein from Bacillus subtilis subsp. subtilis str. 168
To Be Published
|
|
4EXK
 
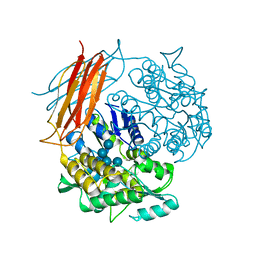 | | A chimera protein containing MBP fused to the C-terminal domain of the uncharacterized protein STM14_2015 from Salmonella enterica | | Descriptor: | Maltose-binding periplasmic protein, uncharacterized protein chimera, TRIETHYLENE GLYCOL, ... | | Authors: | Nocek, B, Hatzos-Skintges, C, Jedrzejczak, R, Babnigg, G, Brown, R.N, Cort, J.R, Heffron, F, Nakayasu, E.S, Adkins, J.N, Joachimiak, A, Program for the Characterization of Secreted Effector Proteins (PCSEP), Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2012-04-30 | | Release date: | 2012-08-08 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (1.28 Å) | | Cite: | A chimera protein containing MBP fused to the C-terminal domain of the uncharacterized protein STM14_2015 form Salmonella enterica
To be Published
|
|
4ICH
 
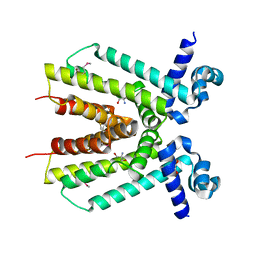 | | Crystal structure of a putative TetR family transcriptional regulator from Saccharomonospora viridis DSM 43017 | | Descriptor: | 1,2-ETHANEDIOL, 2-[3-(2-HYDROXY-1,1-DIHYDROXYMETHYL-ETHYLAMINO)-PROPYLAMINO]-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, BETA-MERCAPTOETHANOL, ... | | Authors: | Filippova, E.V, Minasov, G, Shuvalova, L, Kiryukhina, O, Jedrzejczak, R, Joachimiak, A, Anderson, W.F, Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2012-12-10 | | Release date: | 2013-01-02 | | Last modified: | 2025-03-26 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | Crystal structure of a putative TetR family transcriptional regulator from Saccharomonospora viridis DSM 43017
To be Published
|
|
4JJT
 
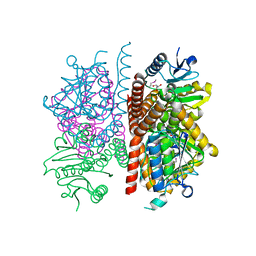 | | The crystal structure of enoyl-CoA hydratase from Mycobacterium tuberculosis H37Rv | | Descriptor: | ACETATE ION, Enoyl-CoA hydratase, GLYCEROL | | Authors: | Tan, K, Holowicki, J, Endres, M, Kim, C.-Y, Kim, H, Hung, L.-W, Terwilliger, T.C, Joachimiak, A, Midwest Center for Structural Genomics (MCSG), Structures of Mtb Proteins Conferring Susceptibility to Known Mtb Inhibitors (MTBI) | | Deposit date: | 2013-03-08 | | Release date: | 2013-03-27 | | Last modified: | 2024-11-27 | | Method: | X-RAY DIFFRACTION (2.496 Å) | | Cite: | The crystal structure of enoyl-CoA hydratase from Mycobacterium tuberculosis H37Rv
To be Published
|
|
4IPT
 
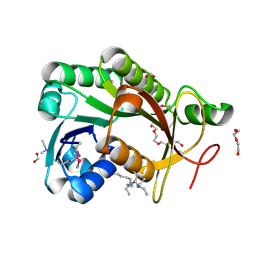 | | The crystal structure of a short-chain dehydrogenases/reductase (ethylated) from Veillonella parvula DSM 2008 | | Descriptor: | CHLORIDE ION, DI(HYDROXYETHYL)ETHER, FORMIC ACID, ... | | Authors: | Tan, K, Hatzos-Skintges, C, Clancy, S, Joachimiak, A, Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2013-01-10 | | Release date: | 2013-02-06 | | Last modified: | 2025-03-26 | | Method: | X-RAY DIFFRACTION (1.546 Å) | | Cite: | The crystal structure of a short-chain dehydrogenases/reductase (ethylated) from Veillonella parvula DSM 2008
To be Published
|
|
4YYF
 
 | | The crystal structure of a glycosyl hydrolase of GH3 family member from [Mycobacterium smegmatis str. MC2 155 | | Descriptor: | ACETATE ION, Beta-N-acetylhexosaminidase, FORMIC ACID, ... | | Authors: | Tan, K, Hatzos-Skintges, C, Clancy, S, Joachimiak, A, Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2015-03-23 | | Release date: | 2015-04-08 | | Last modified: | 2024-11-13 | | Method: | X-RAY DIFFRACTION (1.92 Å) | | Cite: | The crystal structure of a glycosyl hydrolase of GH3 family member from [Mycobacterium smegmatis str. MC2 155
To Be Published
|
|
4GYU
 
 | | Crystal Structure of New Delhi Metallo-beta-Lactamase-1 A121F mutant from Klebsiella pneumoniae | | Descriptor: | Beta-lactamase NDM-1, DI(HYDROXYETHYL)ETHER, GLYCEROL, ... | | Authors: | Kim, Y, Tesar, C, Jedrzejczak, R, Babnigg, J, Binkowski, T.A, Mire, J, Sacchettini, J, Joachimiak, A, Midwest Center for Structural Genomics (MCSG), Structures of Mtb Proteins Conferring Susceptibility to Known Mtb Inhibitors (MTBI) | | Deposit date: | 2012-09-05 | | Release date: | 2012-09-26 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (1.803 Å) | | Cite: | Crystal Structure of New Delhi Metallo-beta-Lactamase-1 A121F mutant from Klebsiella pneumoniae
To be Published, 2012
|
|
4H0D
 
 | | New Delhi Metallo-beta-Lactamase-1 Complexed with Mn from Klebsiella pneumoniae | | Descriptor: | (2R,4S)-2-[(R)-{[(2R)-2-amino-2-phenylacetyl]amino}(carboxy)methyl]-5,5-dimethyl-1,3-thiazolidine-4-carboxylic acid, 1,2-ETHANEDIOL, Beta-lactamase NDM-1, ... | | Authors: | Kim, Y, Tesar, C, Jedrzejczak, R, Babnigg, J, Binkowski, T.A, Mire, J, Sacchettini, J, Joachimiak, A, Midwest Center for Structural Genomics (MCSG), Structures of Mtb Proteins Conferring Susceptibility to Known Mtb Inhibitors (MTBI) | | Deposit date: | 2012-09-07 | | Release date: | 2012-09-26 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (1.498 Å) | | Cite: | New Delhi Metallo-beta-Lactamase-1 Complexed with Mn from Klebsiella pneumoniae
To be Published
|
|
4HCJ
 
 | | Crystal Structure of ThiJ/PfpI Domain Protein from Brachyspira murdochii | | Descriptor: | CHLORIDE ION, FORMIC ACID, MAGNESIUM ION, ... | | Authors: | Kim, Y, Bigelow, L, Jedrzejczak, R, Joachimiak, A, Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2012-09-30 | | Release date: | 2012-10-24 | | Last modified: | 2024-10-09 | | Method: | X-RAY DIFFRACTION (1.12 Å) | | Cite: | Crystal Structure of ThiJ/PfpI Domain Protein from Brachyspira murdochii
To be Published
|
|
1XBW
 
 | | 1.9A Crystal Structure of the protein isdG from Staphylococcus aureus aureus, Structural genomics, MCSG | | Descriptor: | hypothetical protein isdG | | Authors: | Zhang, R, Wu, R, Joachimiak, G, Schneewind, O, Joachimiak, A, Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2004-08-31 | | Release date: | 2004-10-12 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Staphylococcus aureus IsdG and IsdI, heme-degrading enzymes with structural similarity to monooxygenases.
J.Biol.Chem., 280, 2005
|
|
4HBZ
 
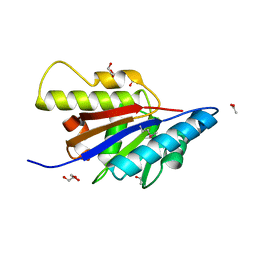 | | The Structure of Putative Phosphohistidine Phosphatase SixA from Nakamurella multipartitia. | | Descriptor: | ACETIC ACID, GLYCEROL, Putative phosphohistidine phosphatase, ... | | Authors: | Cuff, M.E, Holowicki, J, Endres, M, Joachimiak, A, Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2012-09-28 | | Release date: | 2012-10-17 | | Last modified: | 2024-10-09 | | Method: | X-RAY DIFFRACTION (1.55 Å) | | Cite: | The Structure of Putative Phosphohistidine Phosphatase SixA from Nakamurella multipartitia.
TO BE PUBLISHED
|
|
4GMD
 
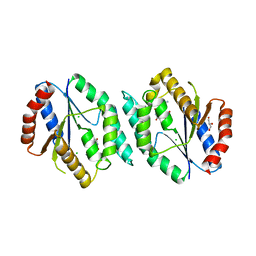 | | The crystal structure of thymidylate kinase from Pseudomonas aeruginosa PAO1 in complex with AZT Monophosphate | | Descriptor: | 3'-AZIDO-3'-DEOXYTHYMIDINE-5'-MONOPHOSPHATE, CALCIUM ION, CHLORIDE ION, ... | | Authors: | Tan, K, Joachimiak, G, Jedrzejczak, R, Sacchettini, J, Joachimiak, A, Midwest Center for Structural Genomics (MCSG), Structures of Mtb Proteins Conferring Susceptibility to Known Mtb Inhibitors (MTBI) | | Deposit date: | 2012-08-15 | | Release date: | 2012-08-29 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (1.98 Å) | | Cite: | The crystal structure of thymidylate kinase from Pseudomonas aeruginosa PAO1 in complex with AZT Monophosphate
To be Published
|
|
4GCN
 
 | | N-terminal domain of stress-induced protein-1 (STI-1) from C.elegans | | Descriptor: | Protein STI-1, TRIETHYLENE GLYCOL | | Authors: | Osipiuk, J, Bigelow, L, Gu, M, Van Oosten-Hawle, P, Morimoto, R.I, Joachimiak, A, Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2012-07-30 | | Release date: | 2012-08-15 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | N-terminal domain of stress-induced protein-1 (STI-1) from C.elegans
To be Published
|
|
4K08
 
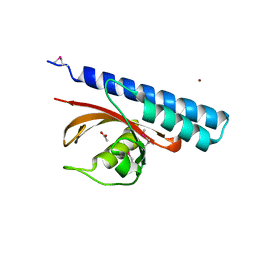 | | Periplasmic sensor domain of chemotaxis protein, Adeh_3718 | | Descriptor: | ACETATE ION, Methyl-accepting chemotaxis sensory transducer, ZINC ION | | Authors: | Pokkuluri, P.R, Mack, J.C, Bearden, J, Rakowski, E, Schiffer, M, Joachimiak, A, Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2013-04-03 | | Release date: | 2013-07-17 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Analysis of periplasmic sensor domains from Anaeromyxobacter dehalogenans 2CP-C: structure of one sensor domain from a histidine kinase and another from a chemotaxis protein.
Microbiologyopen, 2, 2013
|
|
4IQ0
 
 | | Crystal structure of oxidoreductase, Gfo/Idh/MocA family from Streptococcus pneumoniae with reductive methylated Lysine | | Descriptor: | Oxidoreductase, Gfo/Idh/MocA family, POTASSIUM ION | | Authors: | Chang, C, Hatzos-Skintges, C, Abdullah, J, Joachimiak, A, Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2013-01-10 | | Release date: | 2013-01-23 | | Last modified: | 2025-03-26 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Crystal structure of oxidoreductase, Gfo/Idh/MocA family from Streptococcus pneumoniae with reductive methylated Lysine
TO BE PUBLISHED
|
|
4RNL
 
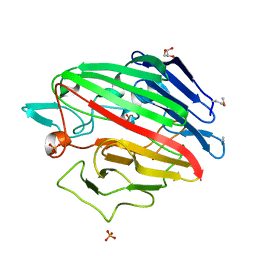 | | The crystal structure of a possible galactose mutarotase from Streptomyces platensis subsp. rosaceus | | Descriptor: | GLYCEROL, PHOSPHATE ION, possible galactose mutarotase | | Authors: | Tan, K, Li, H, Endres, M, Phillips Jr, G.N, Joachimiak, A, Midwest Center for Structural Genomics (MCSG), Enzyme Discovery for Natural Product Biosynthesis (NatPro) | | Deposit date: | 2014-10-24 | | Release date: | 2014-11-26 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | The crystal structure of a possible galactose mutarotase from Streptomyces platensis subsp. rosaceus
To be Published
|
|
4QA9
 
 | | Ensemble refinement of an epoxide hydrolase from Streptomyces carzinostaticus subsp. neocarzinostaticus. | | Descriptor: | 1,2-ETHANEDIOL, Epoxide hydrolase, SULFATE ION | | Authors: | Wang, F, Tan, K, Bigelow, L, Clancy, S, Babnigg, G, Bingman, C.A, Yennamalli, R, Lohman, J, Ma, M, Shen, B, Joachimiak, A, Phillips Jr, G.N, Midwest Center for Structural Genomics (MCSG), Enzyme Discovery for Natural Product Biosynthesis (NatPro) | | Deposit date: | 2014-05-02 | | Release date: | 2014-05-21 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (1.56 Å) | | Cite: | Ensemble refinement of an epoxide hydrolase from Streptomyces carzinostaticus subsp. neocarzinostaticus.
To be Published
|
|
4KQC
 
 | | ABC transporter, LacI family transcriptional regulator from Brachyspira murdochii | | Descriptor: | NITRATE ION, Periplasmic binding protein/LacI transcriptional regulator | | Authors: | Osipiuk, J, Joachimiak, G, Clancy, S, Joachimiak, A, Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2013-05-14 | | Release date: | 2013-05-29 | | Last modified: | 2024-10-09 | | Method: | X-RAY DIFFRACTION (1.62 Å) | | Cite: | ABC transporter, LacI family transcriptional regulator from Brachyspira murdochii
To be Published
|
|
4KSN
 
 | |
4JGQ
 
 | | The crystal structure of sporulation kinase D mutant sensor domain, r131a, from Bacillus subtilis subsp in co-crystallization with pyruvate | | Descriptor: | ACETIC ACID, Sporulation kinase D | | Authors: | Wu, R, Schiffer, M, Gu, M, Joachimiak, A, Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2013-03-01 | | Release date: | 2013-05-15 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (2.63 Å) | | Cite: | Insight into the sporulation phosphorelay: Crystal structure of the sensor domain of Bacillus subtilis histidine kinase, KinD.
Protein Sci., 22, 2013
|
|
4I76
 
 | | Crystal structure of transcriptional regulator TM1030 with octanol | | Descriptor: | 1,2-ETHANEDIOL, OCTAN-1-OL, Transcriptional regulator, ... | | Authors: | Koclega, K.D, Chruszcz, M, Cooper, D.R, Petkowski, J.J, Tkaczuk, K.L, Joachimiak, A, Minor, W, Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2012-11-30 | | Release date: | 2013-01-02 | | Last modified: | 2024-11-27 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Crystal structure of transcriptional regulator TM1030 with octanol
To be Published
|
|
5C00
 
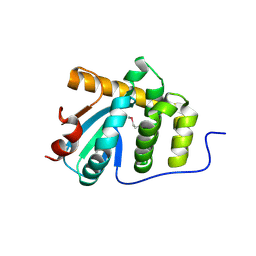 | | MdbA protein, a thiol-disulfide oxidoreductase from Corynebacterium diphtheriae | | Descriptor: | MdbA protein | | Authors: | OSIPIUK, J, REARDON-ROBINSON, M.E, TON-THAT, H, JOACHIMIAK, A, Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2015-06-11 | | Release date: | 2015-07-15 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (1.77 Å) | | Cite: | A thiol-disulfide oxidoreductase of the Gram-positive pathogen Corynebacterium diphtheriae is essential for viability, pilus assembly, toxin production and virulence.
Mol.Microbiol., 98, 2015
|
|
4DBX
 
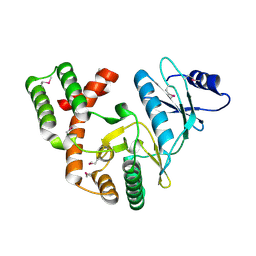 | | Crystal structure of aminoglycoside phosphotransferase APH(2")-ID/APH(2")-IVA | | Descriptor: | APH(2")-ID | | Authors: | Stogios, P.J, Minasov, G, Tan, K, Nocek, B, Singer, A.U, Evdokimova, E, Egorova, E, Di Leo, R, Li, H, Shakya, T, Wright, G.D, Savchenko, A, Anderson, W.F, Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2012-01-16 | | Release date: | 2012-02-01 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (2.004 Å) | | Cite: | A small molecule discrimination map of the antibiotic resistance kinome.
Chem.Biol., 18, 2011
|
|
5C0P
 
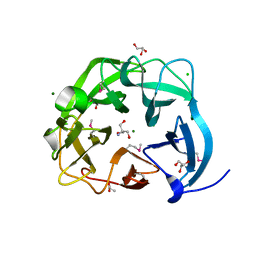 | | The crystal structure of endo-arabinase from Bacteroides thetaiotaomicron VPI-5482 | | Descriptor: | 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, CHLORIDE ION, Endo-arabinase, ... | | Authors: | Tan, K, Cuff, M, Joachimiak, G, Endres, M, Joachimiak, A, Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2015-06-12 | | Release date: | 2015-07-01 | | Last modified: | 2024-11-20 | | Method: | X-RAY DIFFRACTION (1.532 Å) | | Cite: | The crystal structure of endo-arabinase from Bacteroides thetaiotaomicron VPI-5482
To Be Published
|
|
5CD2
 
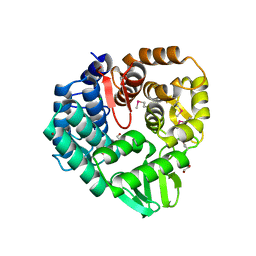 | | The crystal structure of endo-1,4-D-glucanase from Vibrio fischeri ES114 | | Descriptor: | CHLORIDE ION, Endo-1,4-D-glucanase, GLYCEROL, ... | | Authors: | Tan, K, Li, H, Endres, M, Joachimiak, A, Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2015-07-02 | | Release date: | 2015-07-22 | | Last modified: | 2024-11-13 | | Method: | X-RAY DIFFRACTION (1.55 Å) | | Cite: | The crystal structure of endo-1,4-D-glucanase from Vibrio fischeri ES114
To Be Published
|
|
