5CUQ
 
 | | Identification and characterization of novel broad spectrum inhibitors of the flavivirus methyltransferase | | Descriptor: | N,N'-BIS(4-AMINO-2-METHYLQUINOLIN-6-YL)UREA, Nonstructural protein NS5 | | Authors: | Brecher, B, Chen, H, Li, Z, Banavali, N.K, Jones, S.A, Zhang, J, Kramer, L.D, Li, H.M. | | Deposit date: | 2015-07-24 | | Release date: | 2016-02-03 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (1.696 Å) | | Cite: | Identification and Characterization of Novel Broad-Spectrum Inhibitors of the Flavivirus Methyltransferase.
Acs Infect Dis., 1, 2015
|
|
7TQB
 
 | |
7TQA
 
 | | Crystal Structure of monoclonal S9.6 Fab | | Descriptor: | Fab S9.6 heavy chain, Fab S9.6 light chain, GLYCEROL, ... | | Authors: | Bou-Nader, C, Zhang, J. | | Deposit date: | 2022-01-26 | | Release date: | 2022-03-30 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (2.328 Å) | | Cite: | Structural basis of R-loop recognition by the S9.6 monoclonal antibody.
Nat Commun, 13, 2022
|
|
2JXW
 
 | |
2LKZ
 
 | | Solution structure of the second RRM domain of RBM5 | | Descriptor: | RNA-binding protein 5 | | Authors: | Song, Z, Wu, P, Zhang, J, Wu, J, Shi, Y. | | Deposit date: | 2011-10-23 | | Release date: | 2012-08-08 | | Last modified: | 2024-05-29 | | Method: | SOLUTION NMR | | Cite: | Solution structure of the second RRM domain of RBM5 and its unusual binding characters for different RNA targets
Biochemistry, 2012
|
|
4MSU
 
 | | Human GKRP bound to AMG-6861 and Sorbitol-6-phosphate | | Descriptor: | 1,1,1,3,3,3-hexafluoro-2-{4-[4-(thiophen-2-ylsulfonyl)piperazin-1-yl]phenyl}propan-2-ol, D-SORBITOL-6-PHOSPHATE, GLYCEROL, ... | | Authors: | Ashton, K.S, Andrews, K.L, Bryan, M.C, Chen, J, Chen, K, Chen, M, Chmait, S, Croghan, M, Cupples, R, Fotsch, C, Helmering, J, Jordan, S.R, Kurzeja, R.J, Michelsen, K, Pennington, L.D, Poon, S.F, Sivits, G, Van, G, Vonderfecht, S.L, Wahl, R.C, Zhang, J, Lloyd, D.J, Hale, C, St Jean, D.J. | | Deposit date: | 2013-09-18 | | Release date: | 2014-03-12 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Small Molecule Disruptors of the Glucokinase-Glucokinase Regulatory Protein Interaction: 1. Discovery of a Novel Tool Compound for in Vivo Proof-of-Concept.
J.Med.Chem., 57, 2014
|
|
2L43
 
 | | Structural basis for histone code recognition by BRPF2-PHD1 finger | | Descriptor: | Histone H3.3,LINKER,Bromodomain-containing protein 1, ZINC ION | | Authors: | Qin, S, Zhang, J, Wu, J, Shi, Y. | | Deposit date: | 2010-10-01 | | Release date: | 2011-08-31 | | Last modified: | 2024-05-01 | | Method: | SOLUTION NMR | | Cite: | Recognition of unmodified histone H3 by the first PHD finger of Bromodomain-PHD finger protein 2 provides insights into the regulation of histone acetyltransferases MOZ and MORF
To be Published
|
|
2KXJ
 
 | | Solution structure of UBX domain of human UBXD2 protein | | Descriptor: | UBX domain-containing protein 4 | | Authors: | Wu, Q, Huang, H, Zhang, J, Hu, Q, Wu, J, Shi, Y. | | Deposit date: | 2010-05-06 | | Release date: | 2011-05-18 | | Last modified: | 2024-05-01 | | Method: | SOLUTION NMR | | Cite: | Solution strcture of UBX domain of human UBXD2 protein
To be Published
|
|
5HRD
 
 | |
5HRK
 
 | |
8HQI
 
 | |
8HQH
 
 | |
8HQJ
 
 | |
5HJG
 
 | | Crystal Structure of Human Transthyretin in Complex with Tetrabromobisphenol A (TBBPA) | | Descriptor: | 4,4'-propane-2,2-diylbis(2,6-dibromophenol), SODIUM ION, Transthyretin | | Authors: | Begum, A, Iakovleva, I, Brannstrom, K, Zhang, J, Andersson, P, Olofsson, A, Sauer-Eriksson, A.E. | | Deposit date: | 2016-01-13 | | Release date: | 2016-05-04 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (1.4 Å) | | Cite: | Tetrabromobisphenol A Is an Efficient Stabilizer of the Transthyretin Tetramer.
Plos One, 11, 2016
|
|
5HRF
 
 | |
8HVL
 
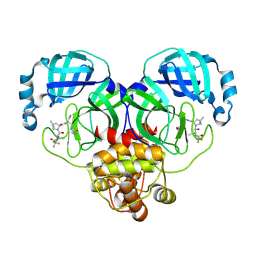 | | Crystal structure of SARS-Cov-2 main protease M49I mutant in complex with PF07321332 | | Descriptor: | (1R,2S,5S)-N-{(1E,2S)-1-imino-3-[(3S)-2-oxopyrrolidin-3-yl]propan-2-yl}-6,6-dimethyl-3-[3-methyl-N-(trifluoroacetyl)-L-valyl]-3-azabicyclo[3.1.0]hexane-2-carboxamide, 3C-like proteinase nsp5 | | Authors: | Zou, X.F, Zhang, J, Li, J. | | Deposit date: | 2022-12-27 | | Release date: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (1.45 Å) | | Cite: | Crystal structure of SARS-Cov-2 main protease
M49I mutant in complex with PF07321332
To Be Published
|
|
8HVO
 
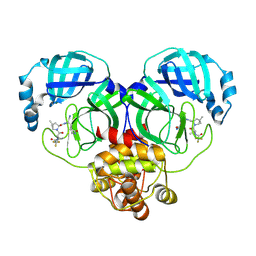 | | Crystal structure of SARS-Cov-2 main protease V186F mutant in complex with PF07321332 | | Descriptor: | (1R,2S,5S)-N-{(1E,2S)-1-imino-3-[(3S)-2-oxopyrrolidin-3-yl]propan-2-yl}-6,6-dimethyl-3-[3-methyl-N-(trifluoroacetyl)-L-valyl]-3-azabicyclo[3.1.0]hexane-2-carboxamide, 3C-like proteinase nsp5 | | Authors: | Zou, X.F, Zhang, J, Li, J. | | Deposit date: | 2022-12-27 | | Release date: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (1.65 Å) | | Cite: | Crystal structure of SARS-Cov-2 main protease
V186F mutant in complex with PF07321332
To Be Published
|
|
8HVN
 
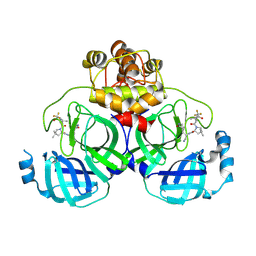 | | Crystal structure of SARS-Cov-2 main protease P132H mutant in complex with PF07321332 | | Descriptor: | (1R,2S,5S)-N-{(1E,2S)-1-imino-3-[(3S)-2-oxopyrrolidin-3-yl]propan-2-yl}-6,6-dimethyl-3-[3-methyl-N-(trifluoroacetyl)-L-valyl]-3-azabicyclo[3.1.0]hexane-2-carboxamide, 3C-like proteinase nsp5 | | Authors: | Li, W.W, Zhang, J, Li, J. | | Deposit date: | 2022-12-27 | | Release date: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Crystal structure of SARS-Cov-2 main protease
P132H mutant in complex with PF07321332
To Be Published
|
|
8HVK
 
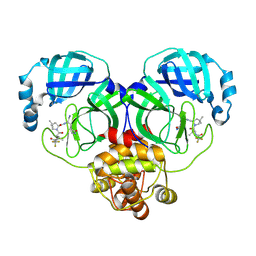 | | Crystal structure of SARS-Cov-2 main protease G15S mutant in complex with PF07321332 | | Descriptor: | (1R,2S,5S)-N-{(1E,2S)-1-imino-3-[(3S)-2-oxopyrrolidin-3-yl]propan-2-yl}-6,6-dimethyl-3-[3-methyl-N-(trifluoroacetyl)-L-valyl]-3-azabicyclo[3.1.0]hexane-2-carboxamide, 3C-like proteinase nsp5 | | Authors: | Zeng, P, Zhang, J, Li, J. | | Deposit date: | 2022-12-27 | | Release date: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (1.63 Å) | | Cite: | Crystal structure of SARS-Cov-2 main protease
G15S mutant in complex with PF07321332
To Be Published
|
|
5HML
 
 | | Crystal Structure of T5 D15 Protein Co-crystallized with Metal Ions | | Descriptor: | 1,2-ETHANEDIOL, 2-[3-(2-HYDROXY-1,1-DIHYDROXYMETHYL-ETHYLAMINO)-PROPYLAMINO]-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, CHLORIDE ION, ... | | Authors: | Flemming, C.S, Feng, M, Sedelnikova, S.E, Zhang, J, Rafferty, J.B, Sayers, J.R, Artymiuk, P.J. | | Deposit date: | 2016-01-16 | | Release date: | 2016-06-01 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (1.482 Å) | | Cite: | Direct observation of DNA threading in flap endonuclease complexes.
Nat.Struct.Mol.Biol., 23, 2016
|
|
5HRE
 
 | | The crystal structure of AsfvPolX:DNA3 binary complex | | Descriptor: | DNA (5'-D(P*AP*GP*GP*AP*TP*CP*CP*T)-3'), DNA polymerase beta-like protein, FORMIC ACID, ... | | Authors: | Chen, Y.Q, Zhang, J, Gan, J.H. | | Deposit date: | 2016-01-23 | | Release date: | 2017-01-18 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (1.75 Å) | | Cite: | The crystal structure of Se-AsfvPolX(L52/163M mutant) in complex with 1nt-gap DNA1
To Be Published
|
|
5HRL
 
 | |
8HZR
 
 | | Crystal structure of SARS-Cov-2 main protease S46F mutant in complex with PF07321332 | | Descriptor: | (1R,2S,5S)-N-{(1E,2S)-1-imino-3-[(3S)-2-oxopyrrolidin-3-yl]propan-2-yl}-6,6-dimethyl-3-[3-methyl-N-(trifluoroacetyl)-L-valyl]-3-azabicyclo[3.1.0]hexane-2-carboxamide, 3C-like proteinase nsp5 | | Authors: | Zeng, X.Y, Zhang, J, Li, J. | | Deposit date: | 2023-01-09 | | Release date: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (1.92 Å) | | Cite: | Crystal structure of SARS-Cov-2 main protease
S46F mutant in complex with PF07321332
To Be Published
|
|
8HQF
 
 | |
5HR9
 
 | |
