1A42
 
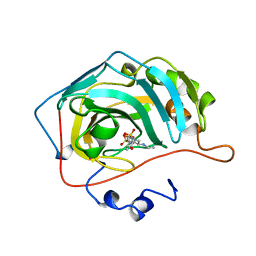 | | HUMAN CARBONIC ANHYDRASE II COMPLEXED WITH BRINZOLAMIDE | | Descriptor: | (4R)-2-(2-ethoxyethyl)-4-(ethylamino)-3,4-dihydro-2H-thieno[3,2-e][1,2]thiazine-6-sulfonamide 1,1-dioxide, CARBONIC ANHYDRASE II, MERCURY (II) ION, ... | | Authors: | Boriack-Sjodin, P.A, Christianson, D.W. | | Deposit date: | 1998-02-10 | | Release date: | 1999-03-23 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (2.25 Å) | | Cite: | Structures of murine carbonic anhydrase IV and human carbonic anhydrase II complexed with brinzolamide: molecular basis of isozyme-drug discrimination.
Protein Sci., 7, 1998
|
|
4YME
 
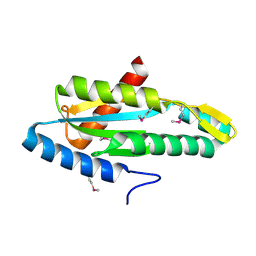 | |
1SHD
 
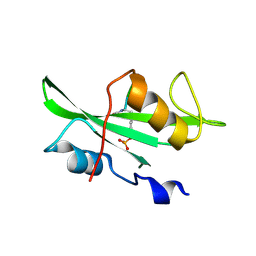 | |
1LHZ
 
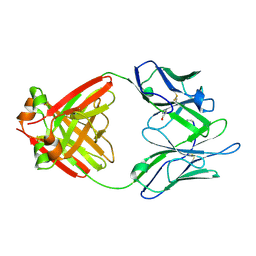 | | Structure of a Human Bence-Jones Dimer Crystallized in U.S. Space Shuttle Mission STS-95: 293K | | Descriptor: | IMMUNOGLOBULIN LAMBDA LIGHT CHAIN | | Authors: | Terzyan, S.S, DeWitt, C.R, Ramsland, P.A, Bourne, P.C, Edmundson, A.B. | | Deposit date: | 2002-04-17 | | Release date: | 2003-07-01 | | Last modified: | 2023-08-16 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Comparison of the three-dimensional structures of a human Bence-Jones dimer
crystallized on Earth and aboard US Space Shuttle Mission STS-95
J.MOL.RECOG., 16, 2003
|
|
1LGV
 
 | | Structure of a Human Bence-Jones Dimer Crystallized in U.S. Space Shuttle Mission STS-95: 100K | | Descriptor: | IMMUNOGLOBULIN LAMBDA LIGHT CHAIN | | Authors: | Terzyan, S.S, DeWitt, C.R, Ramsland, P.A, Bourne, P.C, Edmundson, A.B. | | Deposit date: | 2002-04-16 | | Release date: | 2003-07-01 | | Last modified: | 2023-08-16 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | Comparison of the three-dimensional structures of a human Bence-Jones dimer
crystallized on Earth and aboard US Space Shuttle Mission STS-95
J.MOL.RECOG., 16, 2003
|
|
3FKQ
 
 | |
7V9L
 
 | | Cryo-EM structure of the SV1-Gs complex. | | Descriptor: | GHRH receptor splice variant 1,GHRH receptor splice variant 1,GHRH receptor splice variant 1,SV1, Guanine nucleotide-binding protein G(I)/G(S)/G(O) subunit gamma-2, Guanine nucleotide-binding protein G(I)/G(S)/G(T) subunit beta-1, ... | | Authors: | Cong, Z.T, Zhou, F.L, Zhang, C, Zou, X.Y, Zhang, H.B, Wang, Y.Z, Zhou, Q.T, Cai, X.Q, Liu, Q.F, Li, J, Shao, L.J, Mao, C.Y, Wang, X, Wu, J.H, Xia, T, Zhao, L.H, Jiang, H.L, Zhang, Y, Xu, H.E, Cheng, X, Yang, D.H, Wang, M.W. | | Deposit date: | 2021-08-26 | | Release date: | 2022-04-06 | | Last modified: | 2022-05-04 | | Method: | ELECTRON MICROSCOPY (2.6 Å) | | Cite: | Constitutive signal bias mediated by the human GHRHR splice variant 1.
Proc.Natl.Acad.Sci.USA, 118, 2021
|
|
5SY1
 
 | | Structure of the STRA6 receptor for retinol uptake in complex with calmodulin | | Descriptor: | CALCIUM ION, CHOLESTEROL, Calmodulin, ... | | Authors: | Clarke, O.B, Chen, Y, Mancia, F. | | Deposit date: | 2016-08-10 | | Release date: | 2016-08-24 | | Last modified: | 2024-03-06 | | Method: | ELECTRON MICROSCOPY (3.9 Å) | | Cite: | Structure of the STRA6 receptor for retinol uptake.
Science, 353, 2016
|
|
4JAN
 
 | | crystal structure of broadly neutralizing antibody CH103 in complex with HIV-1 gp120 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, ANTIGEN BINDING FRAGMENT OF HEAVY CHAIN of CH103, ANTIGEN BINDING FRAGMENT OF LIGHT CHAIN of CH103, ... | | Authors: | Zhou, T, Moquin, S, Zheng, A, Srivatsan, S, Kwong, P.D. | | Deposit date: | 2013-02-18 | | Release date: | 2013-04-03 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (3.15 Å) | | Cite: | Co-evolution of a broadly neutralizing HIV-1 antibody and founder virus.
Nature, 496, 2013
|
|
7V9M
 
 | | Cryo-EM structure of the GHRH-bound human GHRHR splice variant 1 complex | | Descriptor: | Guanine nucleotide-binding protein G(I)/G(S)/G(O) subunit gamma-2, Guanine nucleotide-binding protein G(I)/G(S)/G(T) subunit beta-1, Guanine nucleotide-binding protein G(s) subunit alpha isoforms short, ... | | Authors: | Cong, Z.T, Zhou, F.L, Zhang, C, Zou, X.Y, Zhang, H.B, Wang, Y.Z, Zhou, Q.T, Cai, X.Q, Liu, Q.F, Li, J, Shao, L.J, Mao, C.Y, Wang, X, Wu, J.H, Xia, T, Zhao, L.H, Jiang, H.L, Zhang, Y, Xu, H.E, Chen, X, Yang, D.H, Wang, M.W. | | Deposit date: | 2021-08-26 | | Release date: | 2021-10-20 | | Last modified: | 2024-06-19 | | Method: | ELECTRON MICROSCOPY (3.29 Å) | | Cite: | Constitutive signal bias mediated by the human GHRHR splice variant 1.
Proc.Natl.Acad.Sci.USA, 118, 2021
|
|
4JAM
 
 | | Crystal structure of broadly neutralizing anti-hiv-1 antibody ch103 | | Descriptor: | 1,2-ETHANEDIOL, ANTIGEN BINDING FRAGMENT OF HEAVY CHAIN of CH103, ANTIGEN BINDING FRAGMENT OF LIGHT CHAIN of CH103, ... | | Authors: | Zhou, T, Moquin, S, Zheng, A, Srivatsan, S, Kwong, P.D. | | Deposit date: | 2013-02-18 | | Release date: | 2013-04-03 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (1.65 Å) | | Cite: | Co-evolution of a broadly neutralizing HIV-1 antibody and founder virus.
Nature, 496, 2013
|
|
2I0O
 
 | | Crystal structure of Anopheles gambiae Ser/Thr phosphatase complexed with Zn2+ | | Descriptor: | Ser/Thr phosphatase, ZINC ION | | Authors: | Jin, X, Sauder, J.M, Burley, S.K, Shapiro, L, New York SGX Research Center for Structural Genomics (NYSGXRC) | | Deposit date: | 2006-08-10 | | Release date: | 2006-10-24 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Structural genomics of protein phosphatases.
J.STRUCT.FUNCT.GENOM., 8, 2007
|
|
7R73
 
 | |
7RI1
 
 | |
7RI2
 
 | |
7R74
 
 | |
2HY3
 
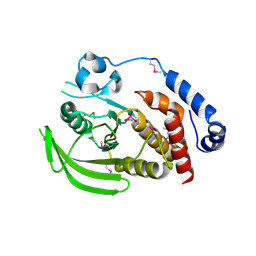 | | Crystal structure of the human tyrosine receptor phosphate gamma in complex with vanadate | | Descriptor: | Receptor-type tyrosine-protein phosphatase gamma, VANADATE ION | | Authors: | Jin, X, Min, T, Bera, A, Mu, H, Sauder, J.M, Freeman, J.C, Reyes, C, Smith, D, Wasserman, S.R, Burley, S.K, Shapiro, L, New York SGX Research Center for Structural Genomics (NYSGXRC) | | Deposit date: | 2006-08-04 | | Release date: | 2006-09-05 | | Last modified: | 2021-02-03 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Structural genomics of protein phosphatases.
J.STRUCT.FUNCT.GENOM., 8, 2007
|
|
7XA9
 
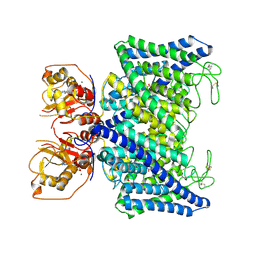 | | Structure of Arabidopsis thaliana CLCa | | Descriptor: | ADENOSINE-5'-TRIPHOSPHATE, Chloride channel protein CLC-a, MAGNESIUM ION, ... | | Authors: | Ji, S, Jin, H, Kaiming, Z, Mingxing, W, Shanshan, L, Long, C. | | Deposit date: | 2022-03-17 | | Release date: | 2023-03-22 | | Method: | ELECTRON MICROSCOPY (2.84 Å) | | Cite: | Cryo-EM structure of the plant nitrate transporter AtCLCa reveals characteristics of the anion-binding site and the ATP-binding pocket.
J.Biol.Chem., 299, 2023
|
|
5WDF
 
 | |
7E0B
 
 | |
4IDJ
 
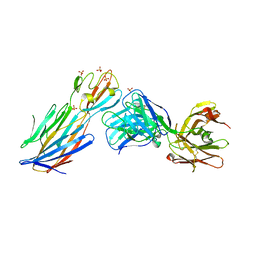 | | S.Aureus a-hemolysin monomer in complex with Fab | | Descriptor: | Alpha-hemolysin, Fab Heavy chain, Fab Light chain, ... | | Authors: | Strop, P. | | Deposit date: | 2012-12-12 | | Release date: | 2013-06-26 | | Last modified: | 2021-05-26 | | Method: | X-RAY DIFFRACTION (3.36 Å) | | Cite: | Mechanism of Action and In Vivo Efficacy of a Human-Derived Antibody against Staphylococcus aureus alpha-Hemolysin.
J.Mol.Biol., 425, 2013
|
|
7TZ0
 
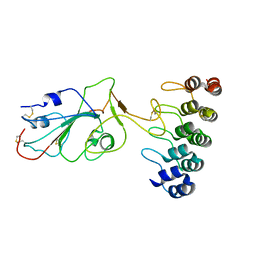 | |
7TYZ
 
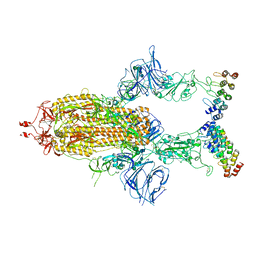 | | Cryo-EM structure of SARS-CoV-2 spike in complex with FSR22, an anti-SARS-CoV-2 DARPin | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, DARPin FSR22, ... | | Authors: | Kwon, Y.D, Gorman, J, Kwong, P.D. | | Deposit date: | 2022-02-15 | | Release date: | 2022-12-07 | | Last modified: | 2023-03-15 | | Method: | ELECTRON MICROSCOPY (3.51 Å) | | Cite: | A potent and broad neutralization of SARS-CoV-2 variants of concern by DARPins.
Nat.Chem.Biol., 19, 2023
|
|
7WOF
 
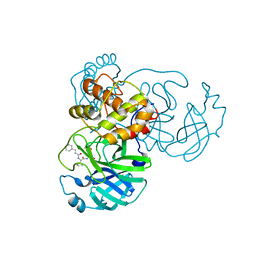 | | SARS-CoV-2 3CLpro | | Descriptor: | (2S,3S)-3-methyl-N-[(2S)-1-oxidanylidene-3-[(3S)-2-oxidanylidenepiperidin-3-yl]propan-2-yl]-2-[[(E)-3-phenylprop-2-enoyl]amino]pentanamide, 3C-like proteinase | | Authors: | Wang, Y, Ye, S. | | Deposit date: | 2022-01-21 | | Release date: | 2022-04-13 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (1.72 Å) | | Cite: | Discovery of SARS-CoV-2 3CL Pro Peptidomimetic Inhibitors through the Catalytic Dyad Histidine-Specific Protein-Ligand Interactions.
Int J Mol Sci, 23, 2022
|
|
7WOH
 
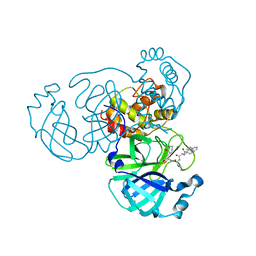 | | SARS-CoV-2 3CLpro | | Descriptor: | (2S)-4-methyl-N-[(2S)-1-oxidanylidene-3-[(3S)-2-oxidanylidenepiperidin-3-yl]propan-2-yl]-2-[[(2S)-3-phenyl-2-[[(E)-3-phenylprop-2-enoyl]amino]propanoyl]amino]pentanamide, 3C-like proteinase | | Authors: | Wang, Y, Ye, S. | | Deposit date: | 2022-01-21 | | Release date: | 2022-04-13 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (1.72 Å) | | Cite: | Discovery of SARS-CoV-2 3CL Pro Peptidomimetic Inhibitors through the Catalytic Dyad Histidine-Specific Protein-Ligand Interactions.
Int J Mol Sci, 23, 2022
|
|
