8SZ7
 
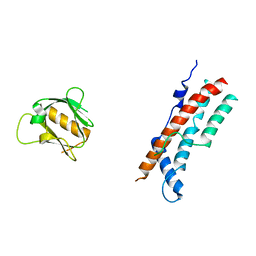 | |
8SZ4
 
 | |
8T0K
 
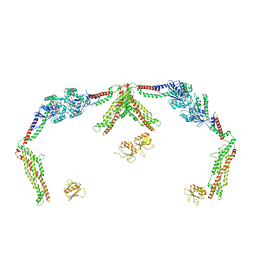 | |
8SXZ
 
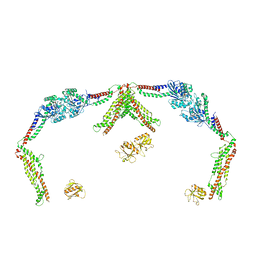 | |
8T0R
 
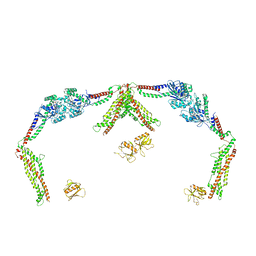 | |
4D8D
 
 | |
2MOM
 
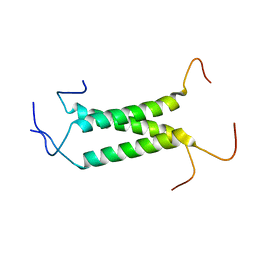 | | Structural insights of TM domain of LAMP-2A in DPC micelles | | Descriptor: | Lysosome-associated membrane glycoprotein 2 | | Authors: | Tjandra, N, Rout, A. | | Deposit date: | 2014-04-27 | | Release date: | 2014-10-29 | | Last modified: | 2024-05-01 | | Method: | SOLUTION NMR | | Cite: | Structure of Transmembrane Domain of Lysosome-associated Membrane Protein Type 2a (LAMP-2A) Reveals Key Features for Substrate Specificity in Chaperone-mediated Autophagy.
J.Biol.Chem., 289, 2014
|
|
2MOF
 
 | | Structural insights of TM domain of LAMP-2A in DPC micelles | | Descriptor: | Lysosome-associated membrane glycoprotein 2 | | Authors: | Tjandra, N, Rout, A. | | Deposit date: | 2014-04-25 | | Release date: | 2014-10-29 | | Last modified: | 2024-05-01 | | Method: | SOLUTION NMR | | Cite: | Structure of Transmembrane Domain of Lysosome-associated Membrane Protein Type 2a (LAMP-2A) Reveals Key Features for Substrate Specificity in Chaperone-mediated Autophagy.
J.Biol.Chem., 289, 2014
|
|
1QA5
 
 | |
1QA4
 
 | |
