7KZZ
 
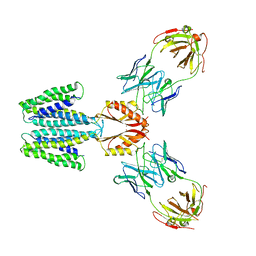 | | Cryo-EM structure of YiiP-Fab complex in Holo state | | Descriptor: | Cadmium and zinc efflux pump FieF, Fab2R heavy chain, Fab2R light chain, ... | | Authors: | Lopez-Redondo, M.L, Fan, S, Koide, A, Koide, S, Beckstein, O, Stokes, D.L. | | Deposit date: | 2020-12-10 | | Release date: | 2021-01-06 | | Last modified: | 2021-07-28 | | Method: | ELECTRON MICROSCOPY (3.42 Å) | | Cite: | Zinc binding alters the conformational dynamics and drives the transport cycle of the cation diffusion facilitator YiiP.
J.Gen.Physiol., 153, 2021
|
|
7KZX
 
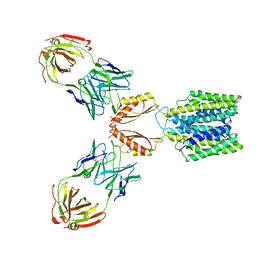 | | Cryo-EM structure of YiiP-Fab complex in Apo state | | Descriptor: | Cadmium and zinc efflux pump FieF, Fab2R heavy chain, Fab2R light chain | | Authors: | Lopez-Redondo, M.L, Fan, S, Koide, A, Koide, S, Beckstein, O, Stokes, D.L. | | Deposit date: | 2020-12-10 | | Release date: | 2021-01-06 | | Last modified: | 2021-07-28 | | Method: | ELECTRON MICROSCOPY (4 Å) | | Cite: | Zinc binding alters the conformational dynamics and drives the transport cycle of the cation diffusion facilitator YiiP.
J.Gen.Physiol., 153, 2021
|
|
7LC3
 
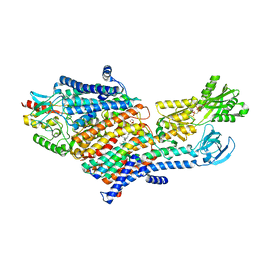 | | CryoEM Structure of KdpFABC in E1-ATP state | | Descriptor: | (2R)-3-(((2-aminoethoxy)(hydroxy)phosphoryl)oxy)-2-(palmitoyloxy)propyl (E)-octadec-9-enoate, MAGNESIUM ION, PHOSPHOMETHYLPHOSPHONIC ACID ADENYLATE ESTER, ... | | Authors: | Sweet, M.E, Larsen, C, Pedersen, B.P, Stokes, D.L. | | Deposit date: | 2021-01-09 | | Release date: | 2021-01-27 | | Last modified: | 2024-05-29 | | Method: | ELECTRON MICROSCOPY (3.23 Å) | | Cite: | Structural basis for potassium transport in prokaryotes by KdpFABC.
Proc.Natl.Acad.Sci.USA, 118, 2021
|
|
7LC6
 
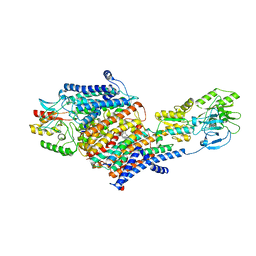 | | Cryo-EM Structure of KdpFABC in E2-P state with BeF3 | | Descriptor: | (2R)-3-(((2-aminoethoxy)(hydroxy)phosphoryl)oxy)-2-(palmitoyloxy)propyl (E)-octadec-9-enoate, MAGNESIUM ION, POTASSIUM ION, ... | | Authors: | Sweet, M.E, Larsen, C, Pedersen, B.P, Stokes, D.L. | | Deposit date: | 2021-01-09 | | Release date: | 2021-01-27 | | Last modified: | 2024-05-29 | | Method: | ELECTRON MICROSCOPY (3.7 Å) | | Cite: | Structural basis for potassium transport in prokaryotes by KdpFABC.
Proc.Natl.Acad.Sci.USA, 118, 2021
|
|
4IFQ
 
 | | Crystal structure of Saccharomyces cerevisiae NUP192, residues 2 to 960 [ScNup192(2-960)] | | Descriptor: | IODIDE ION, Nucleoporin NUP192, SULFATE ION | | Authors: | Sampathkumar, P, Almo, S.C, New York Structural Genomics Research Consortium (NYSGRC), Nucleocytoplasmic Transport: a Target for Cellular Control (NPCXstals) | | Deposit date: | 2012-12-14 | | Release date: | 2013-02-20 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (3.25 Å) | | Cite: | Structure, dynamics, evolution, and function of a major scaffold component in the nuclear pore complex.
Structure, 21, 2013
|
|
8QEE
 
 | | S. cerevisia Niemann-Pick type C protein NCR1 in Peptidisc at pH 7.5 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, CHOLESTEROL HEMISUCCINATE, ... | | Authors: | Frain, K.M, Dedic, E, Nel, L, Olesen, E, Stokes, D, Panyella Pedersen, B. | | Deposit date: | 2023-08-31 | | Release date: | 2023-10-18 | | Last modified: | 2024-04-17 | | Method: | ELECTRON MICROSCOPY (2.43 Å) | | Cite: | Conformational changes in the Niemann-Pick type C1 protein NCR1 drive sterol translocation.
Proc.Natl.Acad.Sci.USA, 121, 2024
|
|
8QED
 
 | | S. cerevisia Niemann-Pick type C protein NCR1 in LMNG at pH 5.5 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, ERGOSTEROL, NPC intracellular sterol transporter 1-related protein 1, ... | | Authors: | Frain, K.M, Nel, L, Dedic, E, Olesen, E, Stokes, D, Panyella Pedersen, B. | | Deposit date: | 2023-08-31 | | Release date: | 2023-10-18 | | Last modified: | 2024-04-17 | | Method: | ELECTRON MICROSCOPY (3.27 Å) | | Cite: | Conformational changes in the Niemann-Pick type C1 protein NCR1 drive sterol translocation.
Proc.Natl.Acad.Sci.USA, 121, 2024
|
|
8QEC
 
 | | S. cerevisia Niemann-Pick type C protein NCR1 in GDN at pH 5.5 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, 2-{[(4-O-alpha-D-glucopyranosyl-alpha-D-glucopyranosyl)oxy]methyl}-4-{[(3beta,9beta,14beta,17beta,25R)-spirost-5-en-3-yl]oxy}butyl 4-O-alpha-D-glucopyranosyl-alpha-D-glucopyranoside, CHOLESTEROL HEMISUCCINATE, ... | | Authors: | Frain, K.M, Dedic, E, Nel, L, Olesen, E, Stokes, D, Panyella Pedersen, B. | | Deposit date: | 2023-08-31 | | Release date: | 2023-10-18 | | Last modified: | 2024-04-17 | | Method: | ELECTRON MICROSCOPY (3.3 Å) | | Cite: | Conformational changes in the Niemann-Pick type C1 protein NCR1 drive sterol translocation.
Proc.Natl.Acad.Sci.USA, 121, 2024
|
|
8QEB
 
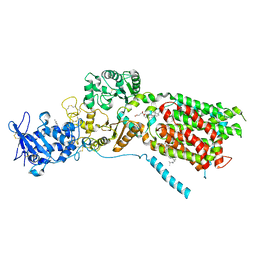 | | S. cerevisia Niemann-Pick type C protein NCR1 in GDN at pH 7.5 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, CHOLESTEROL HEMISUCCINATE, ERGOSTEROL, ... | | Authors: | Frain, K.M, Dedic, E, Nel, L, Olesen, E, Stokes, D, Panyella Pedersen, B. | | Deposit date: | 2023-08-31 | | Release date: | 2023-10-18 | | Last modified: | 2024-04-17 | | Method: | ELECTRON MICROSCOPY (3.3 Å) | | Cite: | Conformational changes in the Niemann-Pick type C1 protein NCR1 drive sterol translocation.
Proc.Natl.Acad.Sci.USA, 121, 2024
|
|
6OL1
 
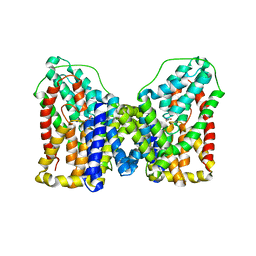 | |
6OL0
 
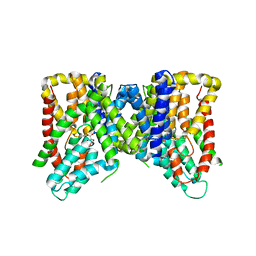 | | Structure of VcINDY bound to Malate | | Descriptor: | (2S)-2-hydroxybutanedioic acid, SODIUM ION, Transporter, ... | | Authors: | Sauer, D.B, Marden, J.J, Wang, D.N. | | Deposit date: | 2019-04-15 | | Release date: | 2020-10-28 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (3.502 Å) | | Cite: | Elevator mechanism dynamics in a sodium-coupled dicarboxylate transporter
Biorxiv, 2022
|
|
6OKZ
 
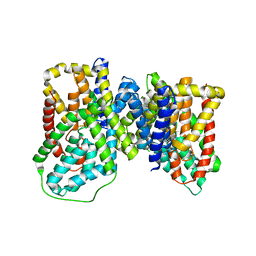 | |
5TVZ
 
 | | Solution NMR structure of Saccharomyces cerevisiae Pom152 Ig-like repeat, residues 718-820 | | Descriptor: | Nucleoporin POM152 | | Authors: | Dutta, K, Sampathkumar, P, Cowburn, D, Almo, S.C, Rout, M.P, Fernandez-Martinez, J. | | Deposit date: | 2016-11-10 | | Release date: | 2017-02-22 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | Molecular Architecture of the Major Membrane Ring Component of the Nuclear Pore Complex.
Structure, 25, 2017
|
|
