3QLT
 
 | |
3QLU
 
 | |
3U64
 
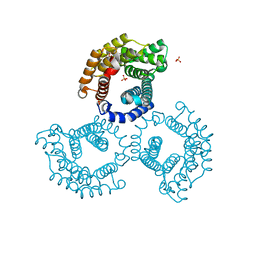 | | The Crystal Structure of Tat-T (Tp0956) | | Descriptor: | Protein TP_0956, SULFATE ION | | Authors: | Tomchick, D.R, Brautigam, C.A, Deka, R.K, Norgard, M.V. | | Deposit date: | 2011-10-12 | | Release date: | 2012-02-22 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Structural, Bioinformatic, and In Vivo Analyses of Two Treponema pallidum Lipoproteins Reveal a Unique TRAP Transporter.
J.Mol.Biol., 416, 2012
|
|
3G3F
 
 | |
3G3K
 
 | |
3G3J
 
 | |
3G3G
 
 | |
3G3H
 
 | |
3G3I
 
 | |
1QO3
 
 | |
