5Z6D
 
 | |
5Z6E
 
 | |
1WBL
 
 | | WINGED BEAN LECTIN COMPLEXED WITH METHYL-ALPHA-D-GALACTOSE | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, CALCIUM ION, MANGANESE (II) ION, ... | | Authors: | Prabu, M.M, Sankaranarayanan, R, Puri, K.D, Sharma, V, Surolia, A, Vijayan, M, Suguna, K. | | Deposit date: | 1997-04-04 | | Release date: | 1998-04-08 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Carbohydrate specificity and quaternary association in basic winged bean lectin: X-ray analysis of the lectin at 2.5 A resolution.
J.Mol.Biol., 276, 1998
|
|
5HT8
 
 | | Crystal structure of clostrillin double mutant (S17H,S19H) in complex with nickel | | Descriptor: | Beta and gamma crystallin, NICKEL (II) ION | | Authors: | Jamkhindikar, A, Srivastava, S.S, Sankaranarayanan, R. | | Deposit date: | 2016-01-26 | | Release date: | 2017-02-01 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.01 Å) | | Cite: | A Transition Metal-Binding, Trimeric beta gamma-Crystallin from Methane-Producing Thermophilic Archaea, Methanosaeta thermophila
Biochemistry, 56, 2017
|
|
5HT7
 
 | |
1Y2Q
 
 | |
2ZLU
 
 | | Horse methemoglobin high salt, pH 7.0 (88% relative humidity) | | Descriptor: | Hemoglobin subunit alpha, Hemoglobin subunit beta, PROTOPORPHYRIN IX CONTAINING FE | | Authors: | Kaushal, P.S, Sankaranarayanan, R, Vijayan, M. | | Deposit date: | 2008-04-10 | | Release date: | 2008-06-24 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Water-mediated variability in the structure of relaxed-state haemoglobin
Acta Crystallogr.,Sect.F, 64, 2008
|
|
2ZLX
 
 | | Horse methemoglobin high salt, pH 7.0 (66% relative humidity) | | Descriptor: | Hemoglobin subunit alpha, Hemoglobin subunit beta, PROTOPORPHYRIN IX CONTAINING FE | | Authors: | Kaushal, P.S, Sankaranarayanan, R, Vijayan, M. | | Deposit date: | 2008-04-10 | | Release date: | 2008-06-24 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Water-mediated variability in the structure of relaxed-state haemoglobin
Acta Crystallogr.,Sect.F, 64, 2008
|
|
2ZLW
 
 | | Horse methemoglobin high salt, pH 7.0 (75% relative humidity) | | Descriptor: | Hemoglobin subunit alpha, Hemoglobin subunit beta, PROTOPORPHYRIN IX CONTAINING FE | | Authors: | Kaushal, P.S, Sankaranarayanan, R, Vijayan, M. | | Deposit date: | 2008-04-10 | | Release date: | 2008-06-24 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (2.9 Å) | | Cite: | Water-mediated variability in the structure of relaxed-state haemoglobin
Acta Crystallogr.,Sect.F, 64, 2008
|
|
2ZLT
 
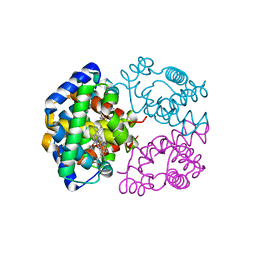 | | Horse methemoglobin high salt, pH 7.0 | | Descriptor: | Hemoglobin subunit alpha, Hemoglobin subunit beta, PROTOPORPHYRIN IX CONTAINING FE | | Authors: | Kaushal, P.S, Sankaranarayanan, R, Vijayan, M. | | Deposit date: | 2008-04-10 | | Release date: | 2008-06-24 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Water-mediated variability in the structure of relaxed-state haemoglobin
Acta Crystallogr.,Sect.F, 64, 2008
|
|
5H4E
 
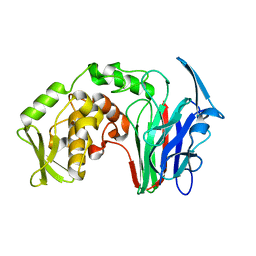 | |
2ZLV
 
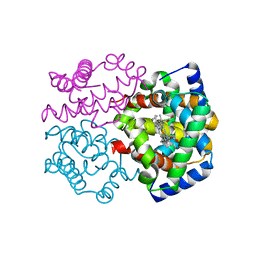 | | Horse methemoglobin high salt, pH 7.0 (79% relative humidity) | | Descriptor: | Hemoglobin subunit alpha, Hemoglobin subunit beta, PROTOPORPHYRIN IX CONTAINING FE | | Authors: | Kaushal, P.S, Sankaranarayanan, R, Vijayan, M. | | Deposit date: | 2008-04-10 | | Release date: | 2008-06-24 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Water-mediated variability in the structure of relaxed-state haemoglobin
Acta Crystallogr.,Sect.F, 64, 2008
|
|
1T2N
 
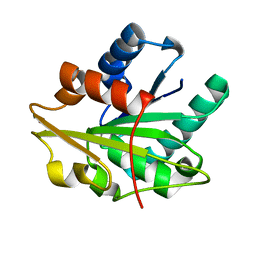 | |
1T4M
 
 | |
2HL0
 
 | | Crystal structure of the editing domain of threonyl-tRNA synthetase from Pyrococcus abyssi in complex with seryl-3'-aminoadenosine | | Descriptor: | SERINE-3'-AMINOADENOSINE, Threonyl-tRNA synthetase | | Authors: | Hussain, T, Kruparani, S.P, Pal, B, Sankaranarayanan, R. | | Deposit date: | 2006-07-06 | | Release date: | 2006-08-29 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (1.86 Å) | | Cite: | Post-transfer editing mechanism of a D-aminoacyl-tRNA deacylase-like domain in threonyl-tRNA synthetase from archaea
Embo J., 25, 2006
|
|
2HL2
 
 | | Crystal structure of the editing domain of threonyl-tRNA synthetase from Pyrococcus abyssi in complex with an analog of seryladenylate | | Descriptor: | 5'-O-(N-(L-SERYL)-SULFAMOYL)ADENOSINE, Threonyl-tRNA synthetase | | Authors: | Hussain, T, Kruparani, S.P, Pal, B, Sankaranarayanan, R. | | Deposit date: | 2006-07-06 | | Release date: | 2006-08-29 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Post-transfer editing mechanism of a D-aminoacyl-tRNA deacylase-like domain in threonyl-tRNA synthetase from archaea
Embo J., 25, 2006
|
|
5ZRN
 
 | |
2HL1
 
 | | Crystal structure of the editing domain of threonyl-tRNA synthetase from Pyrococcus abyssi in complex with seryl-3'-aminoadenosine | | Descriptor: | SERINE-3'-AMINOADENOSINE, Threonyl-tRNA synthetase | | Authors: | Hussain, T, Kruparani, S.P, Pal, B, Sankaranarayanan, R. | | Deposit date: | 2006-07-06 | | Release date: | 2006-08-29 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (2.25 Å) | | Cite: | Post-transfer editing mechanism of a D-aminoacyl-tRNA deacylase-like domain in threonyl-tRNA synthetase from archaea
Embo J., 25, 2006
|
|
2HKZ
 
 | |
2QXU
 
 | |
1NYQ
 
 | | Structure of Staphylococcus aureus threonyl-tRNA synthetase complexed with an analogue of threonyl adenylate | | Descriptor: | 5'-O-(N-(L-THREONYL)-SULFAMOYL)ADENOSINE, ZINC ION, threonyl-tRNA synthetase 1 | | Authors: | Torres-Larios, A, Sankaranarayanan, R, Rees, B, Dock-Bregeon, A.C, Moras, D. | | Deposit date: | 2003-02-13 | | Release date: | 2003-10-28 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (3.2 Å) | | Cite: | Conformational movements and cooperativity upon amino acid, ATP and tRNA binding in threonyl-tRNA synthetase
J.Mol.Biol., 331, 2003
|
|
4DQV
 
 | |
1NYR
 
 | | Structure of Staphylococcus aureus threonyl-tRNA synthetase complexed with ATP | | Descriptor: | ADENOSINE-5'-TRIPHOSPHATE, THREONINE, ZINC ION, ... | | Authors: | Torres-Larios, A, Sankaranarayanan, R, Rees, B, Dock-Bregeon, A.C, Moras, D. | | Deposit date: | 2003-02-13 | | Release date: | 2003-10-28 | | Last modified: | 2023-08-16 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Conformational movements and cooperativity upon amino acid, ATP and tRNA binding in threonyl-tRNA synthetase
J.Mol.Biol., 331, 2003
|
|
1LJ4
 
 | |
1LJI
 
 | |
