1HJU
 
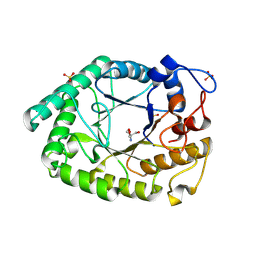 | | Structure of two fungal beta-1,4-galactanases: searching for the basis for temperature and pH optimum. | | Descriptor: | 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, 2-acetamido-2-deoxy-beta-D-glucopyranose, BETA-1,4-GALACTANASE, ... | | Authors: | Le Nours, J, Ryttersgaard, C, Lo Leggio, L, Ostergaard, P.R, Borchert, T.V, Christensen, L.L.H, Larsen, S. | | Deposit date: | 2003-02-27 | | Release date: | 2003-06-02 | | Last modified: | 2020-07-29 | | Method: | X-RAY DIFFRACTION (2.15 Å) | | Cite: | Structure of Two Fungal Beta-1,4-Galactanases: Searching for the Basis for Temperature and Ph Optimum
Protein Sci., 12, 2003
|
|
1HJQ
 
 | | Structure of two fungal beta-1,4-galactanases: searching for the basis for temperature and pH optimum. | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, BETA-1,4-GALACTANASE | | Authors: | Le Nours, J, Ryttersgaard, C, Lo Leggio, L, Ostergaard, P.R, Borchert, T.V, Christensen, L.L.H, Larsen, S. | | Deposit date: | 2003-02-27 | | Release date: | 2003-07-25 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (2.55 Å) | | Cite: | Structure of Two Fungal Beta-1,4-Galactanases: Searching for the Basis for Temperature and Ph Optimum
Protein Sci., 12, 2003
|
|
3ZUD
 
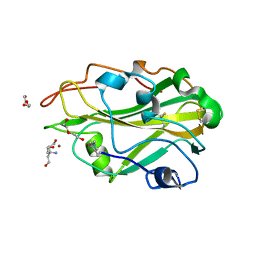 | | THERMOASCUS GH61 ISOZYME A | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, COPPER (II) ION, DI(HYDROXYETHYL)ETHER, ... | | Authors: | Otten, H, Quinlan, R.J, Sweeney, M.D, Poulsen, J.-C.N, Johansen, K.S, Krogh, K.B.R.M, Joergensen, C.I, Tovborg, M, Anthonsen, A, Tryfona, T, Walter, C.P, Dupree, P, Xu, F, Davies, G.J, Walton, P.H, Lo Leggio, L. | | Deposit date: | 2011-07-18 | | Release date: | 2011-09-07 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (1.25 Å) | | Cite: | Insights Into the Oxidative Degradation of Cellulose by a Copper Metalloenzyme that Exploits Biomass Components.
Proc.Natl.Acad.Sci.USA, 108, 2011
|
|
8QAO
 
 | |
7ZE9
 
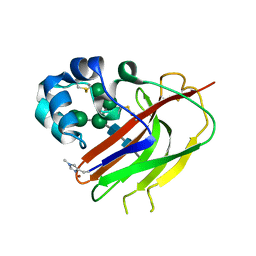 | | Structure of an AA16 LPMO-like protein | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, ACETATE ION, COPPER (II) ION, ... | | Authors: | Huang, Z, Banerjee, S, Muderspach, S.J, Sun, P, van Berkel, W.J.H, Kabel, M.A, Lo Leggio, L. | | Deposit date: | 2022-03-30 | | Release date: | 2023-03-15 | | Last modified: | 2024-05-01 | | Method: | X-RAY DIFFRACTION (2.646 Å) | | Cite: | AA16 Oxidoreductases Boost Cellulose-Active AA9 Lytic Polysaccharide Monooxygenases from Myceliophthora thermophila.
Acs Catalysis, 13, 2023
|
|
2BVY
 
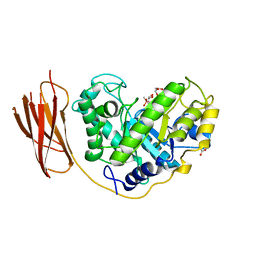 | | The structure and characterization of a modular endo-beta-1,4-mannanase from Cellulomonas fimi | | Descriptor: | ACETATE ION, BETA-1,4-MANNANASE, CACODYLATE ION, ... | | Authors: | Le Nours, J, Anderson, L, Stoll, D, Stalbrand, H, Lo Leggio, L. | | Deposit date: | 2005-07-04 | | Release date: | 2005-09-26 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (2.25 Å) | | Cite: | The Structure and Characterization of a Modular Endo-Beta-1,4-Mannanase from Cellulomonas Fimi
Biochemistry, 44, 2005
|
|
2BVT
 
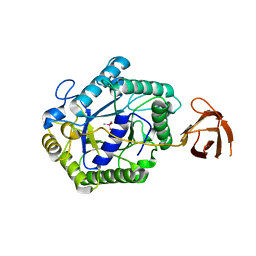 | | The structure of a modular endo-beta-1,4-mannanase from Cellulomonas fimi explains the product specificity of glycoside hydrolase family 26 mannanases. | | Descriptor: | BETA-1,4-MANNANASE, CACODYLATE ION, beta-D-mannopyranose-(1-4)-beta-D-mannopyranose-(1-4)-beta-D-mannopyranose | | Authors: | Le Nours, J, Anderson, L, Stoll, D, Stalbrand, H, Lo Leggio, L. | | Deposit date: | 2005-07-04 | | Release date: | 2005-09-26 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (2.9 Å) | | Cite: | The Structure and Characterization of a Modular Endo-Beta-1,4-Mannanase from Cellulomonas Fimi
Biochemistry, 44, 2005
|
|
4AIE
 
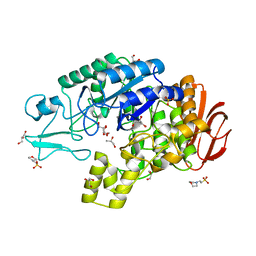 | | Structure of glucan-1,6-alpha-glucosidase from Lactobacillus acidophilus NCFM | | Descriptor: | 2-(N-MORPHOLINO)-ETHANESULFONIC ACID, CALCIUM ION, GLUCAN 1,6-ALPHA-GLUCOSIDASE, ... | | Authors: | Fredslund, F, Navarro Poulsen, J.C, Lo Leggio, L. | | Deposit date: | 2012-02-09 | | Release date: | 2012-08-29 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2.05 Å) | | Cite: | Enzymology and Structure of the Gh13_31 Glucan 1,6-Alpha-Glucosidase that Confers Isomaltooligosaccharide Utilization in the Probiotic Lactobacillus Acidophilus Ncfm.
J.Bacteriol., 194, 2012
|
|
2CCR
 
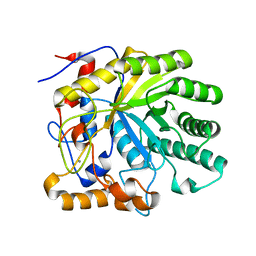 | | Structure of Beta-1,4-Galactanase | | Descriptor: | CALCIUM ION, TRIETHYLENE GLYCOL, YVFO, ... | | Authors: | Le Nours, J, De Maria, L, Welner, D, Jorgensen, C.T, Christensen, L.L.H, Larsen, S, Lo Leggio, L. | | Deposit date: | 2006-01-18 | | Release date: | 2006-03-21 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Investigating the Binding of Beta-1,4-Galactan to Bacillus Licheniformis Beta-1,4-Galactanase by Crystallography and Computational Modeling.
Proteins, 75, 2009
|
|
2GFT
 
 | | Crystal structure of the E263A nucleophile mutant of Bacillus licheniformis endo-beta-1,4-galactanase in complex with galactotriose | | Descriptor: | CALCIUM ION, Glycosyl Hydrolase Family 53, beta-D-galactopyranose-(1-4)-beta-D-galactopyranose-(1-4)-beta-D-galactopyranose | | Authors: | Welner, D, Le Nours, J, De Maria, L, Jorgensen, C.T, Christensen, L.L.H, Larsen, S, Lo Leggio, L. | | Deposit date: | 2006-03-23 | | Release date: | 2006-06-13 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Oligosaccharide binding of Bacillus licheniformis endo-beta-1,4-galactanase
To be Published
|
|
8B48
 
 | | Structure of Lentithecium fluviatile carbohydrate esterase from the CE15 family (LfCE15C) | | Descriptor: | Carbohydrate esterase family 15 protein, FORMIC ACID, alpha-D-mannopyranose-(1-3)-[alpha-D-mannopyranose-(1-6)]beta-D-mannopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Scholzen, K, Mazurkewich, S, Poulsen, J.C.N, Larsbrink, J, Lo Leggio, L. | | Deposit date: | 2022-09-20 | | Release date: | 2023-06-07 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2.65 Å) | | Cite: | Structural and functional investigation of a fungal member of carbohydrate esterase family 15 with potential specificity for rare xylans.
Acta Crystallogr D Struct Biol, 79, 2023
|
|
5JO9
 
 | | Structural characterization of the thermostable Bradyrhizobium japonicum d-sorbitol dehydrogenase | | Descriptor: | PHOSPHATE ION, Ribitol 2-dehydrogenase, sorbitol | | Authors: | Fredslund, F, Otten, H, Navarro Poulsen, J.-C, Lo Leggio, L. | | Deposit date: | 2016-05-02 | | Release date: | 2016-11-09 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (2.894 Å) | | Cite: | Structural characterization of the thermostable Bradyrhizobium japonicumD-sorbitol dehydrogenase.
Acta Crystallogr F Struct Biol Commun, 72, 2016
|
|
5XB7
 
 | | GH42 alpha-L-arabinopyranosidase from Bifidobacterium animalis subsp. lactis Bl-04 | | Descriptor: | Beta-galactosidase, GLYCEROL, SULFATE ION | | Authors: | Viborg, A.H, Katayama, T, Arakawa, T, Abou Hachem, M, Lo Leggio, L, Kitaoka, M, Svensson, B, Fushinobu, S. | | Deposit date: | 2017-03-16 | | Release date: | 2017-11-01 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Discovery of alpha-l-arabinopyranosidases from human gut microbiome expands the diversity within glycoside hydrolase family 42.
J. Biol. Chem., 292, 2017
|
|
1UT7
 
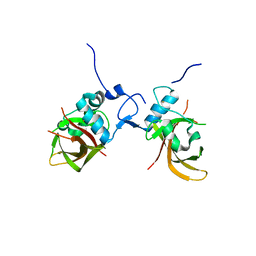 | | Structure of the conserved domain of ANAC, a member of the NAC family of transcription factors | | Descriptor: | GOLD ION, NO APICAL MERISTEM PROTEIN | | Authors: | Ernst, H.A, Olsen, A.N, Skriver, K, Larsen, S, Lo Leggio, L. | | Deposit date: | 2003-12-04 | | Release date: | 2004-03-19 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Structure of the Conserved Domain of Anac, a Member of the Nac Family of Transcription Factors
Embo Rep., 5, 2004
|
|
1UT4
 
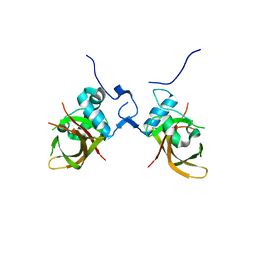 | | Structure of the conserved domain of ANAC, a member of the NAC family of transcription factors | | Descriptor: | NO APICAL MERISTEM PROTEIN | | Authors: | Ernst, H.A, Olsen, A.N, Skriver, K, Larsen, S, Lo Leggio, L. | | Deposit date: | 2003-12-03 | | Release date: | 2004-03-19 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Structure of the Conserved Domain of Anac, a Member of the Nac Family of Transcription Factors
Embo Rep., 5, 2004
|
|
4UR4
 
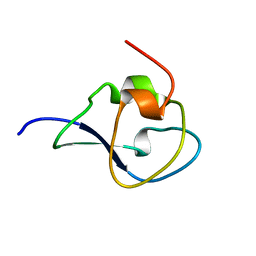 | | Structure of the type III fish antifreeze protein from Zoarces viviparus ZvAFP13 | | Descriptor: | ANTIFREEZE PROTEIN 13 | | Authors: | Wilkens, C, Poulsen, J.-C.N, Ramloev, H, Lo Leggio, L. | | Deposit date: | 2014-06-26 | | Release date: | 2014-07-23 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (1.45 Å) | | Cite: | Purification, Crystal Structure Determination and Functional Characterization of Type III Antifreeze Proteins from the European Eelpout Zoarces Viviparus.
Cryobiology, 69, 2014
|
|
4UR6
 
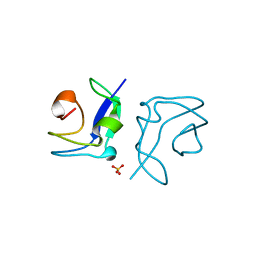 | | Structure of the type III fish antifreeze protein from Zoarces viviparus ZvAFP6 | | Descriptor: | SULFATE ION, TYPE III ANTIFREEZE PROTEIN 6 | | Authors: | Wilkens, C, Poulsen, J.-C.N, Ramloev, H, Lo Leggio, L. | | Deposit date: | 2014-06-26 | | Release date: | 2014-07-23 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (1.2 Å) | | Cite: | Purification, Crystal Structure Determination and Functional Characterization of Type III Antifreeze Proteins from the European Eelpout Zoarces Viviparus.
Cryobiology, 69, 2014
|
|
2G3M
 
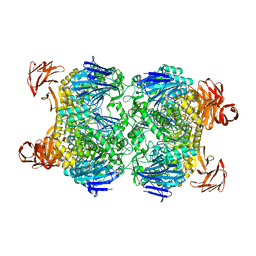 | | Crystal structure of the Sulfolobus solfataricus alpha-glucosidase MalA | | Descriptor: | Alpha-glucosidase | | Authors: | Ernst, H.A, Lo Leggio, L, Willemoes, M, Leonard, G, Blum, P, Larsen, S. | | Deposit date: | 2006-02-20 | | Release date: | 2006-05-02 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (2.55 Å) | | Cite: | Structure of the Sulfolobus solfataricus alpha-Glucosidase: Implications for Domain Conservation and Substrate Recognition in GH31.
J.Mol.Biol., 358, 2006
|
|
2G3N
 
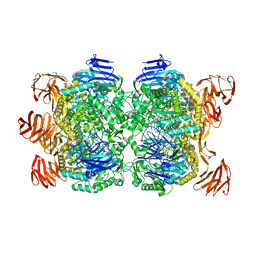 | | Crystal structure of the Sulfolobus solfataricus alpha-glucosidase MalA in complex with beta-octyl-glucopyranoside | | Descriptor: | Alpha-glucosidase, octyl beta-D-glucopyranoside | | Authors: | Ernst, H.A, Lo Leggio, L, Willemoes, M, Leonard, G, Blum, P, Larsen, S. | | Deposit date: | 2006-02-20 | | Release date: | 2006-05-02 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (2.55 Å) | | Cite: | Structure of the Sulfolobus solfataricus alpha-Glucosidase: Implications for Domain Conservation and Substrate Recognition in GH31.
J.Mol.Biol., 358, 2006
|
|
8B4G
 
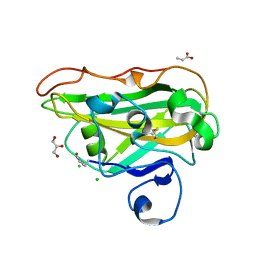 | | Structure of a fungal LPMO bound to ligands | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, ACRYLIC ACID, CHLORIDE ION, ... | | Authors: | Banerjee, S, Huang, Z, Brander, S, Johansen, K.S, Lo Leggio, L. | | Deposit date: | 2022-09-20 | | Release date: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (1.496 Å) | | Cite: | Structure of a fungal LPMO bound to ligands
To Be Published
|
|
3EII
 
 | | Zinc-bound glycoside hydrolase 61 E from Thielavia terrestris | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, GLYCEROL, SULFATE ION, ... | | Authors: | Salbo, R, Welner, D, Lo Leggio, L, Harris, P, McFarland, K. | | Deposit date: | 2008-09-16 | | Release date: | 2009-10-27 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (2.25 Å) | | Cite: | Stimulation of lignocellulosic biomass hydrolysis by proteins of glycoside hydrolase family 61: structure and function of a large, enigmatic family.
Biochemistry, 49, 2010
|
|
3EJA
 
 | | Magnesium-bound glycoside hydrolase 61 isoform E from Thielavia terrestris | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, GLYCEROL, MAGNESIUM ION, ... | | Authors: | Salbo, R, Welner, D, Lo Leggio, L, Harris, P, McFarland, K. | | Deposit date: | 2008-09-18 | | Release date: | 2009-10-27 | | Last modified: | 2020-07-29 | | Method: | X-RAY DIFFRACTION (1.902 Å) | | Cite: | Stimulation of lignocellulosic biomass hydrolysis by proteins of glycoside hydrolase family 61: structure and function of a large, enigmatic family.
Biochemistry, 49, 2010
|
|
8E1W
 
 | | Neutron crystal structure of Panus similis AA9A at room temperature | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, CHLORIDE ION, COPPER (II) ION, ... | | Authors: | Meilleur, F, Tandrup, T, Lo Leggio, L. | | Deposit date: | 2022-08-11 | | Release date: | 2023-01-11 | | Last modified: | 2024-04-03 | | Method: | NEUTRON DIFFRACTION (2.1 Å), X-RAY DIFFRACTION | | Cite: | Joint X-ray/neutron structure of Lentinus similis AA9_A at room temperature.
Acta Crystallogr.,Sect.F, 79, 2023
|
|
6Q3R
 
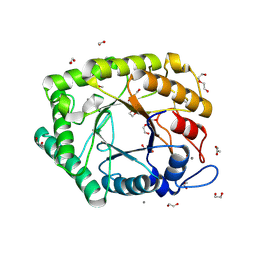 | | ASPERGILLUS ACULEATUS GALACTANASE | | Descriptor: | 1,2-ETHANEDIOL, 1-(2-METHOXY-ETHOXY)-2-{2-[2-(2-METHOXY-ETHOXY]-ETHOXY}-ETHANE, ACETATE ION, ... | | Authors: | Muderspach, S.J, Torpenholt, S, Lo Leggio, L, Poulsen, J.C.N. | | Deposit date: | 2018-12-04 | | Release date: | 2019-06-12 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (2.69 Å) | | Cite: | Structure of Aspergillus aculeatus beta-1,4-galactanase in complex with galactobiose.
Acta Crystallogr.,Sect.F, 75, 2019
|
|
5T7K
 
 | | X-ray crystal structure of AA13 LPMO | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, AoAA13, ZINC ION | | Authors: | Frandsen, K.E.H, Poulsen, J.-C.N, Tovborg, M, Johansen, K.S, Lo Leggio, L. | | Deposit date: | 2016-09-05 | | Release date: | 2017-01-18 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (1.3 Å) | | Cite: | Learning from oligosaccharide soaks of crystals of an AA13 lytic polysaccharide monooxygenase: crystal packing, ligand binding and active-site disorder.
Acta Crystallogr D Struct Biol, 73, 2017
|
|
