1MZM
 
 | |
6KTB
 
 | | Crystal structure of B. halodurans MntR in apo form | | Descriptor: | HTH-type transcriptional regulator MntR, MAGNESIUM ION, MANGANESE (II) ION, ... | | Authors: | Lee, J.Y, Lee, M.Y. | | Deposit date: | 2019-08-26 | | Release date: | 2019-12-04 | | Last modified: | 2024-03-27 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Structural analysis of the manganese transport regulator MntR from Bacillus halodurans in apo and manganese bound forms.
Plos One, 14, 2019
|
|
6KTA
 
 | | Crystal structure of B. halodurans MntR in apo form | | Descriptor: | GLYCEROL, HTH-type transcriptional regulator MntR | | Authors: | Lee, J.Y, Lee, M.Y. | | Deposit date: | 2019-08-26 | | Release date: | 2019-12-04 | | Last modified: | 2024-03-27 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Structural analysis of the manganese transport regulator MntR from Bacillus halodurans in apo and manganese bound forms.
Plos One, 14, 2019
|
|
3N8X
 
 | | Crystal Structure of Cyclooxygenase-1 in Complex with Nimesulide | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, 4-NITRO-2-PHENOXYMETHANESULFONANILIDE, ... | | Authors: | Lee, J.Y. | | Deposit date: | 2010-05-28 | | Release date: | 2010-07-28 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (2.75 Å) | | Cite: | Comparison of Cyclooxygenase-1 Crystal Structures: Cross-Talk between Monomers Comprising Cyclooxygenase-1 Homodimers
Biochemistry, 49, 2010
|
|
2IS2
 
 | | Crystal structure of UvrD-DNA binary complex | | Descriptor: | 33-MER, DNA helicase II, FORMIC ACID, ... | | Authors: | Yang, W, Lee, J.Y. | | Deposit date: | 2006-10-16 | | Release date: | 2007-01-09 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | UvrD helicase unwinds DNA one base pair at a time by a two-part power stroke.
Cell(Cambridge,Mass.), 127, 2006
|
|
2IS1
 
 | | Crystal structure of UvrD-DNA-SO4 complex | | Descriptor: | 5'-D(*CP*GP*AP*GP*CP*AP*CP*TP*GP*C)-3', 5'-D(*GP*CP*AP*GP*TP*GP*CP*TP*CP*GP*TP*TP*TP*TP*TP*TP*T)-3', DNA helicase II, ... | | Authors: | Yang, W, Lee, J.Y. | | Deposit date: | 2006-10-16 | | Release date: | 2007-01-09 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (2.9 Å) | | Cite: | UvrD helicase unwinds DNA one base pair at a time by a two-part power stroke.
Cell(Cambridge,Mass.), 127, 2006
|
|
2IS6
 
 | | Crystal structure of UvrD-DNA-ADPMgF3 ternary complex | | Descriptor: | 5'-D(*CP*GP*AP*GP*CP*AP*CP*TP*GP*CP*AP*GP*TP*GP*CP*TP*CP*GP*TP*TP*GP*TP*TP*AP*T)-3', ADENOSINE-5'-DIPHOSPHATE, DNA helicase II, ... | | Authors: | Yang, W, Lee, J.Y. | | Deposit date: | 2006-10-16 | | Release date: | 2007-01-09 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | UvrD helicase unwinds DNA one base pair at a time by a two-part power stroke.
Cell(Cambridge,Mass.), 127, 2006
|
|
2IS4
 
 | | Crystal structure of UvrD-DNA-ADPNP ternary complex | | Descriptor: | 25-MER, DNA helicase II, MAGNESIUM ION, ... | | Authors: | Yang, W, Lee, J.Y. | | Deposit date: | 2006-10-16 | | Release date: | 2007-01-09 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | UvrD helicase unwinds DNA one base pair at a time by a two-part power stroke.
Cell(Cambridge,Mass.), 127, 2006
|
|
5GPA
 
 | |
5GPC
 
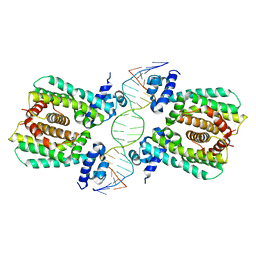 | | Structural analysis of fatty acid degradation regulator FadR from Bacillus halodurans | | Descriptor: | DNA (5'-D(P*CP*AP*TP*GP*AP*AP*TP*GP*AP*GP*TP*AP*TP*TP*CP*AP*TP*TP*CP*AP*T)-3'), DNA (5'-D(P*GP*AP*TP*GP*AP*AP*TP*GP*AP*AP*TP*AP*CP*TP*CP*AP*TP*TP*CP*AP*T)-3'), Transcriptional regulator (TetR/AcrR family) | | Authors: | Lee, J.Y, Yeo, H.K, Park, T.W. | | Deposit date: | 2016-08-01 | | Release date: | 2017-03-01 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Structural basis of operator sites recognition and effector binding in the TetR family transcription regulator FadR.
Nucleic Acids Res., 45, 2017
|
|
5GP9
 
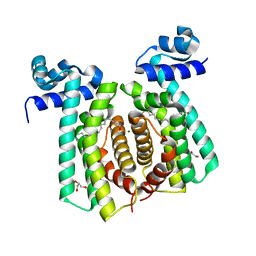 | | Structural analysis of fatty acid degradation regulator FadR from Bacillus halodurans | | Descriptor: | GLYCEROL, MAGNESIUM ION, PALMITIC ACID, ... | | Authors: | Lee, J.Y, Yeo, H.K, Park, Y.W. | | Deposit date: | 2016-08-01 | | Release date: | 2017-03-01 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (1.755 Å) | | Cite: | Structural basis of operator sites recognition and effector binding in the TetR family transcription regulator FadR.
Nucleic Acids Res., 45, 2017
|
|
4O6J
 
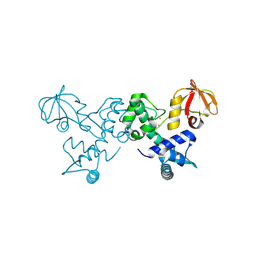 | | Crystal sturucture of T. acidophilum IdeR | | Descriptor: | FE (II) ION, Iron-dependent transcription repressor related protein | | Authors: | Lee, J.Y, Yeo, H.K. | | Deposit date: | 2013-12-20 | | Release date: | 2014-05-21 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Structural analysis and insight into metal-ion activation of the iron-dependent regulator from Thermoplasma acidophilum.
Acta Crystallogr.,Sect.D, 70, 2014
|
|
4O5V
 
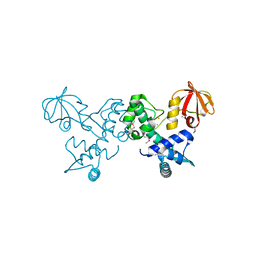 | | Crystal structure of T. acidophilum IdeR | | Descriptor: | FE (II) ION, Iron-dependent transcription repressor related protein | | Authors: | Lee, J.Y, Yeo, H.K. | | Deposit date: | 2013-12-20 | | Release date: | 2014-11-05 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Structural analysis and insight into metal-ion activation of the iron-dependent regulator from Thermoplasma acidophilum.
Acta Crystallogr.,Sect.D, 70, 2014
|
|
8CUB
 
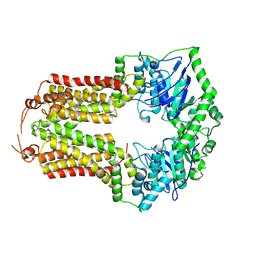 | |
3RYO
 
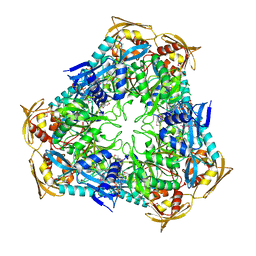 | | Crystal Structure of Enhanced Intracellular Survival (Eis) Protein from Mycobacterium tuberculosis with Acetyl CoA | | Descriptor: | ACETYL COENZYME *A, Enhanced intracellular survival protein | | Authors: | Kim, K.H, An, D.R, Yoon, J.Y, Kim, H.S, Yoon, H.J, Song, J, Im, H.N, Kim, J, Kim, D.J, Lee, S.J, Kim, H.J, Lee, J.Y, Suh, S.W. | | Deposit date: | 2011-05-11 | | Release date: | 2012-05-23 | | Last modified: | 2024-10-09 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Mycobacterium tuberculosis Eis protein initiates suppression of host immune responses by acetylation of DUSP16/MKP-7
Proc.Natl.Acad.Sci.USA, 2012
|
|
3KT1
 
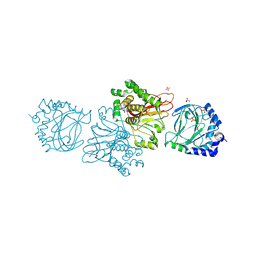 | | Crystal structure of Tpa1 from Saccharomyces cerevisiae, a component of the messenger ribonucleoprotein complex | | Descriptor: | FE (III) ION, GLYCEROL, PKHD-type hydroxylase TPA1, ... | | Authors: | Kim, H.S, Kim, H.L, Kim, K.H, Kim, D.J, Lee, S.J, Yoon, J.Y, Yoon, H.J, Lee, H.Y, Park, S.B, Kim, S.-J, Lee, J.Y, Suh, S.W. | | Deposit date: | 2009-11-24 | | Release date: | 2010-01-19 | | Last modified: | 2017-11-01 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Crystal structure of Tpa1 from Saccharomyces cerevisiae, a component of the messenger ribonucleoprotein complex
Nucleic Acids Res., 38, 2010
|
|
3KT4
 
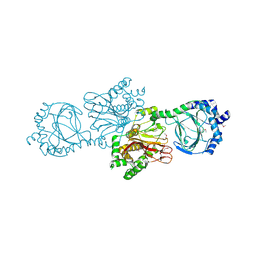 | | Crystal structure of Tpa1 from Saccharomyces cerevisiae, a component of the messenger ribonucleoprotein complex | | Descriptor: | FE (III) ION, PKHD-type hydroxylase TPA1 | | Authors: | Kim, H.S, Kim, H.L, Kim, K.H, Kim, D.J, Lee, S.J, Yoon, J.Y, Yoon, H.J, Lee, H.Y, Park, S.B, Kim, S.-J, Lee, J.Y, Suh, S.W. | | Deposit date: | 2009-11-24 | | Release date: | 2010-01-19 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (2.73 Å) | | Cite: | Crystal structure of Tpa1 from Saccharomyces cerevisiae, a component of the messenger ribonucleoprotein complex
Nucleic Acids Res., 38, 2010
|
|
3KT7
 
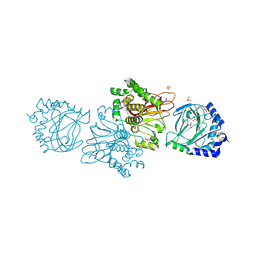 | | Crystal structure of Tpa1 from Saccharomyces cerevisiae, a component of the messenger ribonucleoprotein complex | | Descriptor: | 2-OXOGLUTARIC ACID, FE (III) ION, GLYCEROL, ... | | Authors: | Kim, H.S, Kim, H.L, Kim, K.H, Kim, D.J, Lee, S.J, Yoon, J.Y, Yoon, H.J, Lee, H.Y, Park, S.B, Kim, S.-J, Lee, J.Y, Suh, S.W. | | Deposit date: | 2009-11-24 | | Release date: | 2010-01-19 | | Last modified: | 2017-11-01 | | Method: | X-RAY DIFFRACTION (1.77 Å) | | Cite: | Crystal structure of Tpa1 from Saccharomyces cerevisiae, a component of the messenger ribonucleoprotein complex
Nucleic Acids Res., 38, 2010
|
|
2WH9
 
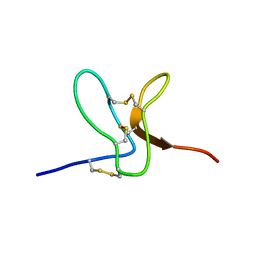 | | Solution structure of GxTX-1E | | Descriptor: | GUANGXITOXIN-1EGXTX-1E | | Authors: | Lee, S.K, Jung, H.H, Lee, J.Y, Lee, C.W, Kim, J.I. | | Deposit date: | 2009-05-02 | | Release date: | 2010-05-26 | | Last modified: | 2023-06-14 | | Method: | SOLUTION NMR | | Cite: | Solution Structure of Gxtx-1E, a High Affinity Tarantula Toxin Interacting with Voltage Sensors in Kv2.1 Potassium Channels.
Biochemistry, 49, 2010
|
|
1C02
 
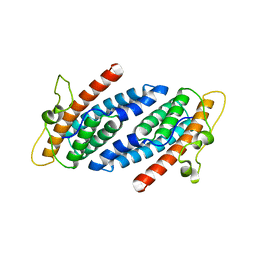 | | CRYSTAL STRUCTURE OF YEAST YPD1P | | Descriptor: | PHOSPHOTRANSFERASE YPD1P | | Authors: | Song, H.K, Lee, J.Y, Lee, M.G, Suh, S.W. | | Deposit date: | 1999-07-14 | | Release date: | 2000-01-15 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Insights into eukaryotic multistep phosphorelay signal transduction revealed by the crystal structure of Ypd1p from Saccharomyces cerevisiae.
J.Mol.Biol., 293, 1999
|
|
1C03
 
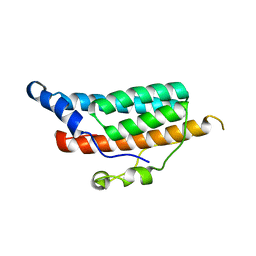 | | CRYSTAL STRUCTURE OF YPD1P (TRICLINIC FORM) | | Descriptor: | HYPOTHETICAL PROTEIN YDL235C | | Authors: | Song, H.K, Lee, J.Y, Lee, M.G, Suh, S.W. | | Deposit date: | 1999-07-14 | | Release date: | 2000-01-15 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Insights into eukaryotic multistep phosphorelay signal transduction revealed by the crystal structure of Ypd1p from Saccharomyces cerevisiae.
J.Mol.Biol., 293, 1999
|
|
1IX1
 
 | | Crystal Structure of P.aeruginosa Peptide deformylase Complexed with Antibiotic Actinonin | | Descriptor: | (CARBAMOYLMETHYL-CARBOXYMETHYL-AMINO)-ACETIC ACID, ACTINONIN, ZINC ION, ... | | Authors: | Kim, H.-W, Yoon, H.-J, Lee, J.Y, Han, B.W, Yang, J.K, Lee, B.I, Ahn, H.J, Lee, H.H, Suh, S.W. | | Deposit date: | 2002-06-07 | | Release date: | 2003-09-09 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | Crystal structure of peptide deformylase from Staphylococcus aureus in complex with actinonin, a naturally occurring antibacterial agent
Proteins, 57, 2004
|
|
4EDB
 
 | | Structures of monomeric hemagglutinin and its complex with an Fab fragment of a neutralizing antibody that binds to H1 subtype influenza viruses: molecular basis of infectivity of 2009 pandemic H1N1 influenza A viruses | | Descriptor: | Hemagglutinin | | Authors: | Kim, K.H, Cho, K.J, Lee, J.H, Park, Y.H, Khan, T.G, Lee, J.Y, Kang, S.H, Alam, I. | | Deposit date: | 2012-03-27 | | Release date: | 2013-05-22 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Insight into structural diversity of influenza virus haemagglutinin
J.Gen.Virol., 94, 2013
|
|
4F15
 
 | | Molecular basis of infectivity of 2009 pandemic H1N1 influenza A viruses | | Descriptor: | Fab fragment, heavy chain, light chain, ... | | Authors: | Kim, K.H, Cho, K.J, Lee, J.H, Park, Y.H, Khan, T.G, Lee, J.Y, Kang, S.H, Alam, I. | | Deposit date: | 2012-05-06 | | Release date: | 2013-05-15 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (2.81 Å) | | Cite: | Molecular basis of infectivity of 2009 pandemic H1N1 influenza A viruses
To be Published
|
|
3SXN
 
 | | Mycobacterium tuberculosis Eis protein initiates modulation of host immune responses by acetylation of DUSP16/MKP-7 | | Descriptor: | COENZYME A, Enhanced intracellular survival protein | | Authors: | Kim, K.H, An, D.R, Yoon, J.Y, Kim, H.S, Yoon, H.J, Song, J.S, Im, H.N, Kim, J, Kim, D.J, Lee, S.J, Kim, H.J, Lee, J.Y, Suh, S.W. | | Deposit date: | 2011-07-15 | | Release date: | 2012-05-23 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (2.03 Å) | | Cite: | Mycobacterium tuberculosis Eis protein initiates suppression of host immune responses by acetylation of DUSP16/MKP-7
Proc.Natl.Acad.Sci.USA, 109, 2012
|
|
