5GWG
 
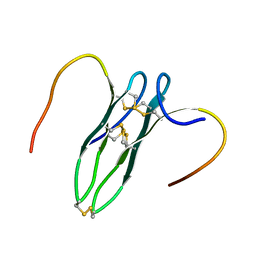 | | Solution structure of rattusin | | Descriptor: | Defensin alpha-related sequence 1 | | Authors: | Lee, C.W, Min, H.J. | | Deposit date: | 2016-09-11 | | Release date: | 2017-04-12 | | Last modified: | 2023-06-14 | | Method: | SOLUTION NMR | | Cite: | Rattusin structure reveals a novel defensin scaffold formed by intermolecular disulfide exchanges
Sci Rep, 7, 2017
|
|
7DVN
 
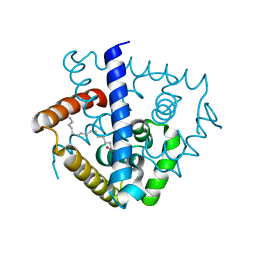 | | Crystal structure of a MarR family protein in complex with a lipid-like effector molecule from the psychrophilic bacterium Paenisporosarcina sp. TG-14 | | Descriptor: | MarR family transcriptional regulator, PALMITIC ACID | | Authors: | Lee, C.W, Hwang, J, Do, H, Lee, J.H. | | Deposit date: | 2021-01-14 | | Release date: | 2021-11-24 | | Last modified: | 2024-05-29 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Crystal structure of a MarR family protein from the psychrophilic bacterium Paenisporosarcina sp. TG-14 in complex with a lipid-like molecule.
Iucrj, 8, 2021
|
|
7C4X
 
 | |
5WT7
 
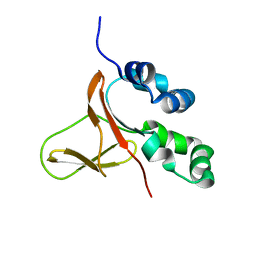 | | FAS1-IV domain of Human Periostin | | Descriptor: | Periostin | | Authors: | Yun, H, Lee, C.W. | | Deposit date: | 2016-12-09 | | Release date: | 2017-12-13 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | 1H,13C, and 15N resonance assignments of FAS1-IV domain of human periostin, a component of extracellular matrix proteins.
Biomol NMR Assign, 12, 2018
|
|
6LVP
 
 | |
4RHE
 
 | | Crystal structure of UbiX, an aromatic acid decarboxylase from the Colwellia psychrerythraea 34H | | Descriptor: | 3-octaprenyl-4-hydroxybenzoate carboxy-lyase, FLAVIN MONONUCLEOTIDE, SULFATE ION | | Authors: | Do, H, Kim, S.J, Lee, C.W, Kim, H.-W, Park, H.H, Kim, H.M, Park, H, Park, H.J, Lee, J.H. | | Deposit date: | 2014-10-02 | | Release date: | 2015-02-18 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (2.003 Å) | | Cite: | Crystal structure of UbiX, an aromatic acid decarboxylase from the psychrophilic bacterium Colwellia psychrerythraea that undergoes FMN-induced conformational changes.
Sci Rep, 5, 2015
|
|
4RHF
 
 | | Crystal structure of UbiX mutant V47S from Colwellia psychrerythraea 34H | | Descriptor: | 3-octaprenyl-4-hydroxybenzoate carboxy-lyase, SULFATE ION | | Authors: | Do, H, Kim, S.J, Lee, C.W, Kim, H.-W, Park, H.H, Kim, H.M, Park, H, Park, H.J, Lee, J.H. | | Deposit date: | 2014-10-02 | | Release date: | 2015-02-18 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (1.764 Å) | | Cite: | Crystal structure of UbiX, an aromatic acid decarboxylase from the psychrophilic bacterium Colwellia psychrerythraea that undergoes FMN-induced conformational changes.
Sci Rep, 5, 2015
|
|
2WH9
 
 | | Solution structure of GxTX-1E | | Descriptor: | GUANGXITOXIN-1EGXTX-1E | | Authors: | Lee, S.K, Jung, H.H, Lee, J.Y, Lee, C.W, Kim, J.I. | | Deposit date: | 2009-05-02 | | Release date: | 2010-05-26 | | Last modified: | 2023-06-14 | | Method: | SOLUTION NMR | | Cite: | Solution Structure of Gxtx-1E, a High Affinity Tarantula Toxin Interacting with Voltage Sensors in Kv2.1 Potassium Channels.
Biochemistry, 49, 2010
|
|
6LVO
 
 | |
4ZZ7
 
 | | Crystal structure of methylmalonate-semialdehyde dehydrogenase (DddC) from Oceanimonas doudoroffii | | Descriptor: | Methylmalonate-semialdehyde dehydrogenase, NICOTINAMIDE-ADENINE-DINUCLEOTIDE | | Authors: | Do, H, Lee, C.W, Lee, S.G, Kang, H, Park, C.M, Kim, H.J, Park, H, Park, H, Lee, J.H. | | Deposit date: | 2015-05-22 | | Release date: | 2016-04-06 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (2.9 Å) | | Cite: | Crystal structure and modeling of the tetrahedral intermediate state of methylmalonate-semialdehyde dehydrogenase (MMSDH) from Oceanimonas doudoroffii.
J. Microbiol., 54, 2016
|
|
7XQG
 
 | | Hemichannel-focused structure of C-terminal truncated connexin43/Cx43/GJA1 gap junction intercellular channel in POPE nanodiscs (GCN conformation) | | Descriptor: | CHOLESTEROL HEMISUCCINATE, Gap junction alpha-1 protein, PHOSPHATIDYLETHANOLAMINE | | Authors: | Lee, H.J, Cha, H.J, Jeong, H, Lee, S.N, Lee, C.W, Woo, J.S. | | Deposit date: | 2022-05-07 | | Release date: | 2023-01-25 | | Last modified: | 2023-08-02 | | Method: | ELECTRON MICROSCOPY (3.8 Å) | | Cite: | Conformational changes in the human Cx43/GJA1 gap junction channel visualized using cryo-EM.
Nat Commun, 14, 2023
|
|
7XQH
 
 | | Hemichannel-focused structure of C-terminal truncated connexin43/Cx43/GJA1 gap junction intercellular channel in POPE nanodiscs (GCN-TM1i conformation) | | Descriptor: | C-terminal deletion mutant of gap junction alpha-1 protein (Cx43-M257) | | Authors: | Lee, H.J, Cha, H.J, Jeong, H, Lee, S.N, Lee, C.W, Woo, J.S. | | Deposit date: | 2022-05-07 | | Release date: | 2023-01-25 | | Last modified: | 2024-10-30 | | Method: | ELECTRON MICROSCOPY (3.8 Å) | | Cite: | Conformational changes in the human Cx43/GJA1 gap junction channel visualized using cryo-EM.
Nat Commun, 14, 2023
|
|
7XQJ
 
 | | Hemichannel-focused structure of C-terminal truncated connexin43/Cx43/GJA1 gap junction intercellular channel in POPE nanodiscs (PLN conformation) | | Descriptor: | Gap junction alpha-1 protein | | Authors: | Lee, H.J, Cha, H.J, Jeong, H, Lee, S.N, Lee, C.W, Woo, J.S. | | Deposit date: | 2022-05-07 | | Release date: | 2023-01-25 | | Last modified: | 2023-05-03 | | Method: | ELECTRON MICROSCOPY (4 Å) | | Cite: | Conformational changes in the human Cx43/GJA1 gap junction channel visualized using cryo-EM.
Nat Commun, 14, 2023
|
|
7XQI
 
 | | Hemichannel-focused structure of C-terminal truncated connexin43/Cx43/GJA1 gap junction intercellular channel in POPE nanodiscs (FIN conformation) | | Descriptor: | Gap junction alpha-1 protein | | Authors: | Lee, H.J, Cha, H.J, Jeong, H, Lee, S.N, Lee, C.W, Woo, J.S. | | Deposit date: | 2022-05-07 | | Release date: | 2023-01-25 | | Last modified: | 2024-10-30 | | Method: | ELECTRON MICROSCOPY (3.7 Å) | | Cite: | Conformational changes in the human Cx43/GJA1 gap junction channel visualized using cryo-EM.
Nat Commun, 14, 2023
|
|
7XQ9
 
 | | Structure of connexin43/Cx43/GJA1 gap junction intercellular channel in GDN detergents at pH ~8.0 | | Descriptor: | Gap junction alpha-1 protein | | Authors: | Lee, H.J, Cha, H.J, Jeong, H, Lee, S.N, Lee, C.W, Woo, J.S. | | Deposit date: | 2022-05-07 | | Release date: | 2023-01-25 | | Last modified: | 2024-10-30 | | Method: | ELECTRON MICROSCOPY (3.3 Å) | | Cite: | Conformational changes in the human Cx43/GJA1 gap junction channel visualized using cryo-EM.
Nat Commun, 14, 2023
|
|
7XQB
 
 | | Structure of connexin43/Cx43/GJA1 gap junction intercellular channel in POPE/CHS nanodiscs at pH ~8.0 | | Descriptor: | CHOLESTEROL HEMISUCCINATE, Gap junction alpha-1 protein, PHOSPHATIDYLETHANOLAMINE, ... | | Authors: | Lee, H.J, Cha, H.J, Jeong, H, Lee, S.N, Lee, C.W, Woo, J.S. | | Deposit date: | 2022-05-07 | | Release date: | 2023-02-01 | | Last modified: | 2024-10-30 | | Method: | ELECTRON MICROSCOPY (3 Å) | | Cite: | Conformational changes in the human Cx43/GJA1 gap junction channel visualized using cryo-EM.
Nat Commun, 14, 2023
|
|
7XQF
 
 | | Structure of C-terminal truncated connexin43/Cx43/GJA1 gap junction intercellular channel in POPE/CHS nanodiscs | | Descriptor: | CHOLESTEROL HEMISUCCINATE, Gap junction alpha-1 protein, PHOSPHATIDYLETHANOLAMINE, ... | | Authors: | Lee, H.J, Cha, H.J, Jeong, H, Lee, S.N, Lee, C.W, Woo, J.S. | | Deposit date: | 2022-05-07 | | Release date: | 2023-02-22 | | Last modified: | 2024-10-16 | | Method: | ELECTRON MICROSCOPY (2.3 Å) | | Cite: | Conformational changes in the human Cx43/GJA1 gap junction channel visualized using cryo-EM.
Nat Commun, 14, 2023
|
|
7XQD
 
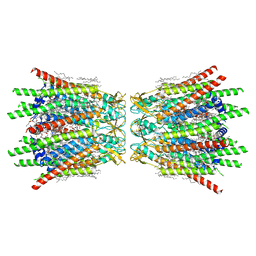 | | Structure of C-terminal truncated connexin43/Cx43/GJA1 gap junction intercellular channel in POPE/CHS nanodiscs (C1 symmetry) | | Descriptor: | CHOLESTEROL HEMISUCCINATE, Gap junction alpha-1 protein, PHOSPHATIDYLETHANOLAMINE, ... | | Authors: | Lee, H.J, Cha, H.J, Jeong, H, Lee, S.N, Lee, C.W, Woo, J.S. | | Deposit date: | 2022-05-07 | | Release date: | 2023-02-22 | | Last modified: | 2023-05-03 | | Method: | ELECTRON MICROSCOPY (2.7 Å) | | Cite: | Conformational changes in the human Cx43/GJA1 gap junction channel visualized using cryo-EM.
Nat Commun, 14, 2023
|
|
7XRH
 
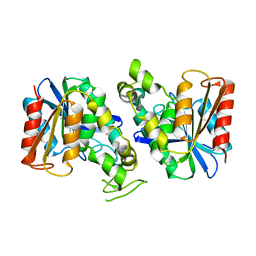 | | Feruloyl esterase from Lactobacillus acidophilus | | Descriptor: | Cinnamoyl esterase | | Authors: | Hwang, J, Lee, C.W, Lee, J.H, Do, H. | | Deposit date: | 2022-05-10 | | Release date: | 2023-05-17 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Feruloyl Esterase ( La Fae) from Lactobacillus acidophilus : Structural Insights and Functional Characterization for Application in Ferulic Acid Production.
Int J Mol Sci, 24, 2023
|
|
7XRI
 
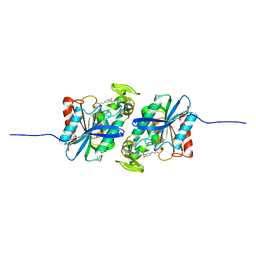 | | Feruloyl esterase mutant -S106A | | Descriptor: | Cinnamoyl esterase, ethyl (2E)-3-(4-hydroxy-3-methoxyphenyl)prop-2-enoate | | Authors: | Hwang, J.S, Lee, J.H, Do, H, Lee, C.W. | | Deposit date: | 2022-05-10 | | Release date: | 2023-05-17 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.19 Å) | | Cite: | Feruloyl Esterase ( La Fae) from Lactobacillus acidophilus : Structural Insights and Functional Characterization for Application in Ferulic Acid Production.
Int J Mol Sci, 24, 2023
|
|
5XGW
 
 | |
1N6A
 
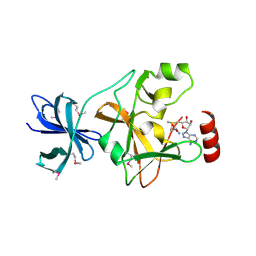 | | Structure of SET7/9 | | Descriptor: | S-ADENOSYLMETHIONINE, SET domain-containing protein 7 | | Authors: | Kwon, T.W, Chang, J.H, Kwak, E, Lee, C.W, Joachimiak, A, Kim, Y.C, Lee, J, Cho, Y. | | Deposit date: | 2002-11-09 | | Release date: | 2003-02-04 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Mechanism of histone lysine methyl transfer revealed by the structure of SET7/9-AdoMet
EMBO J., 22, 2003
|
|
5XGX
 
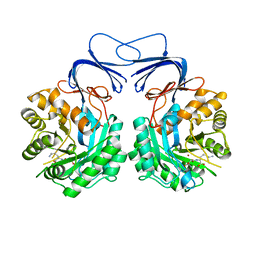 | | Crystal structure of colwellia psychrerythraea strain 34H isoaspartyl dipeptidase E80Q mutant complexed with beta-isoaspartyl lysine | | Descriptor: | D-ASPARTIC ACID, D-LYSINE, Isoaspartyl dipeptidase, ... | | Authors: | Lee, J.H, Lee, C.W, Park, S.H. | | Deposit date: | 2017-04-18 | | Release date: | 2018-02-28 | | Last modified: | 2024-03-27 | | Method: | X-RAY DIFFRACTION (2.33 Å) | | Cite: | Crystal structure and functional characterization of an isoaspartyl dipeptidase (CpsIadA) from Colwellia psychrerythraea strain 34H.
PLoS ONE, 12, 2017
|
|
5YL7
 
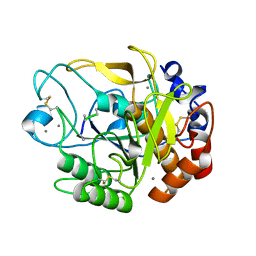 | | Proteases from Pseudoalteromonas arctica PAMC 21717 (Pro21717) | | Descriptor: | CALCIUM ION, Copurified unknown peptide, Pseudoalteromonas arctica PAMC 21717 | | Authors: | Lee, J.H, Lee, C.W. | | Deposit date: | 2017-10-17 | | Release date: | 2018-01-31 | | Last modified: | 2018-09-12 | | Method: | X-RAY DIFFRACTION (1.4 Å) | | Cite: | Crystal structure of a cold-active protease (Pro21717) from the psychrophilic bacterium, Pseudoalteromonas arctica PAMC 21717, at 1.4 angstrom resolution: Structural adaptations to cold and functional analysis of a laundry detergent enzyme
PLoS ONE, 13, 2018
|
|
6AGQ
 
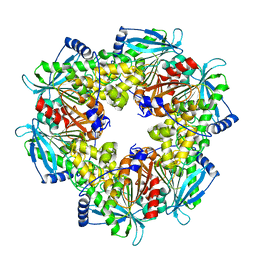 | | Acetyl xylan esterase from Paenibacillus sp. R4 | | Descriptor: | ZINC ION, acetyl xylan esterase | | Authors: | Park, S, Lee, C.W, Lee, J.H. | | Deposit date: | 2018-08-13 | | Release date: | 2018-10-10 | | Last modified: | 2024-03-27 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Crystal structure and functional characterization of a cold-active acetyl xylan esterase (PbAcE) from psychrophilic soil microbe Paenibacillus sp.
PLoS ONE, 13, 2018
|
|
