5VYK
 
 | | Crystal structure of the BRS domain of BRAF in complex with the CC-SAM domain of KSR1 | | Descriptor: | Chimera protein of BRS domain of BRAF and CC-SAM domain of KSR1,Serine/threonine-protein kinase B-raf, GLYCEROL | | Authors: | Maisonneuve, P, Kurinov, I, Marullo, S.A, Lavoie, H, Thevakumaran, N, Sahmi, M, Jin, T, Therrien, M, SIcheri, F. | | Deposit date: | 2017-05-25 | | Release date: | 2018-02-14 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (1.749 Å) | | Cite: | MEK drives BRAF activation through allosteric control of KSR proteins.
Nature, 554, 2018
|
|
6LOZ
 
 | | crystal structure of alpha-momorcharin in complex with adenine | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, ADENINE, Ribosome-inactivating protein momordin I | | Authors: | Fan, X, Jin, T. | | Deposit date: | 2020-01-07 | | Release date: | 2020-11-18 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (1.08 Å) | | Cite: | Atomic-resolution structures of type I ribosome inactivating protein alpha-momorcharin with different substrate analogs.
Int.J.Biol.Macromol., 164, 2020
|
|
6LP0
 
 | | crystal structure of alpha-momorcharin in complex with AMP | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, ADENOSINE MONOPHOSPHATE, Ribosome-inactivating protein momordin I | | Authors: | Fan, X, Jin, T. | | Deposit date: | 2020-01-07 | | Release date: | 2020-11-18 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (1.519 Å) | | Cite: | Atomic-resolution structures of type I ribosome inactivating protein alpha-momorcharin with different substrate analogs.
Int.J.Biol.Macromol., 164, 2020
|
|
6LOR
 
 | | crystal structure of alpha-momorcharin in complex with ADP | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, ADENOSINE-5'-DIPHOSPHATE, Ribosome-inactivating protein momordin I | | Authors: | Fan, X, Jin, T. | | Deposit date: | 2020-01-07 | | Release date: | 2020-11-18 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (1.35 Å) | | Cite: | Atomic-resolution structures of type I ribosome inactivating protein alpha-momorcharin with different substrate analogs.
Int.J.Biol.Macromol., 164, 2020
|
|
6LOW
 
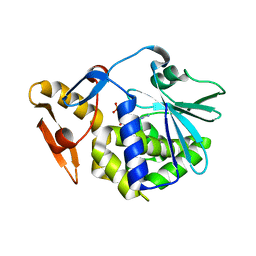 | | crystal structure of alpha-momorcharin in complex with GMP | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, GUANOSINE-5'-MONOPHOSPHATE, Ribosome-inactivating protein momordin I | | Authors: | Fan, X, Jin, T. | | Deposit date: | 2020-01-07 | | Release date: | 2020-11-18 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (1.39 Å) | | Cite: | Atomic-resolution structures of type I ribosome inactivating protein alpha-momorcharin with different substrate analogs.
Int.J.Biol.Macromol., 164, 2020
|
|
6LOQ
 
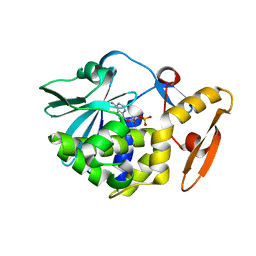 | | crystal structure of alpha-momorcharin in complex with cAMP | | Descriptor: | ADENOSINE-3',5'-CYCLIC-MONOPHOSPHATE, Ribosome-inactivating protein momordin I | | Authors: | Fan, X, Jin, T. | | Deposit date: | 2020-01-07 | | Release date: | 2020-11-18 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (1.331 Å) | | Cite: | Atomic-resolution structures of type I ribosome inactivating protein alpha-momorcharin with different substrate analogs.
Int.J.Biol.Macromol., 164, 2020
|
|
6LOY
 
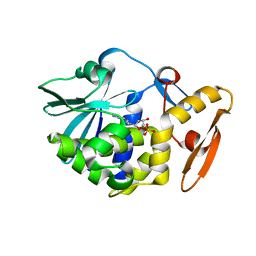 | | crystal structure of alpha-momorcharin in complex with dAMP | | Descriptor: | 2'-DEOXYADENOSINE-5'-MONOPHOSPHATE, Ribosome-inactivating protein momordin I | | Authors: | Fan, X, Jin, T. | | Deposit date: | 2020-01-07 | | Release date: | 2020-11-18 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (1.35 Å) | | Cite: | Atomic-resolution structures of type I ribosome inactivating protein alpha-momorcharin with different substrate analogs.
Int.J.Biol.Macromol., 164, 2020
|
|
6LOV
 
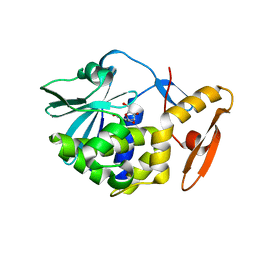 | | crystal structure of alpha-momorcharin in complex with xanthosine | | Descriptor: | 2,3-dihydroxanthosine, Ribosome-inactivating protein momordin I | | Authors: | Fan, X, Jin, T. | | Deposit date: | 2020-01-07 | | Release date: | 2020-11-18 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (1.35 Å) | | Cite: | Atomic-resolution structures of type I ribosome inactivating protein alpha-momorcharin with different substrate analogs.
Int.J.Biol.Macromol., 164, 2020
|
|
5GP1
 
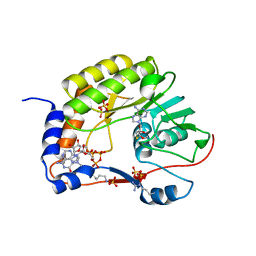 | | Crystal structure of ZIKV NS5 Methyltransferase in complex with GTP and SAH | | Descriptor: | NICKEL (II) ION, P1-7-METHYLGUANOSINE-P3-ADENOSINE-5',5'-TRIPHOSPHATE, RNA-directed RNA polymerase NS5, ... | | Authors: | Zhang, C, Jin, T. | | Deposit date: | 2016-07-30 | | Release date: | 2016-12-07 | | Last modified: | 2017-10-18 | | Method: | X-RAY DIFFRACTION (2.444 Å) | | Cite: | Structure of the NS5 methyltransferase from Zika virus and implications in inhibitor design
Biochem. Biophys. Res. Commun., 492, 2017
|
|
5GOZ
 
 | |
5YGS
 
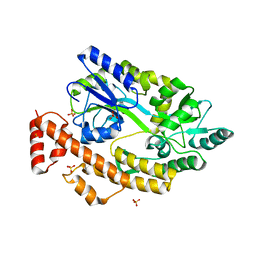 | | Human TNFRSF25 death domain | | Descriptor: | Human TNRSF25 death domain, SULFATE ION, alpha-D-glucopyranose-(1-4)-alpha-D-glucopyranose-(1-4)-alpha-D-glucopyranose | | Authors: | Yin, X, Jin, T. | | Deposit date: | 2017-09-26 | | Release date: | 2018-10-03 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.691 Å) | | Cite: | Crystal structure and activation mechanism of DR3 death domain.
Febs J., 286, 2019
|
|
5ZTC
 
 | |
6JOX
 
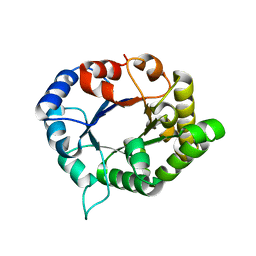 | | triosephosphate isomerase-scylla paramamosain | | Descriptor: | Triosephosphate isomerase | | Authors: | Xia, F, Jin, T. | | Deposit date: | 2019-03-25 | | Release date: | 2019-12-04 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (1.803 Å) | | Cite: | Crystal Structure Analysis and Conformational Epitope Mutation of Triosephosphate Isomerase, a Mud Crab Allergen.
J.Agric.Food Chem., 67, 2019
|
|
6JYM
 
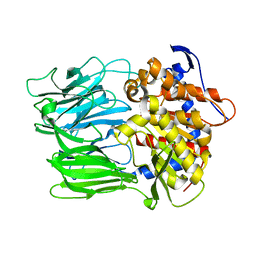 | |
5YEV
 
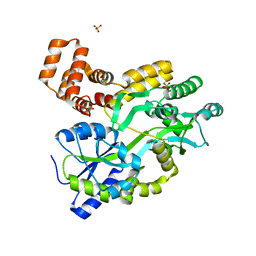 | | Murine DR3 death domain | | Descriptor: | SULFATE ION, TNFRSF25 death domain | | Authors: | Yin, X, Jin, T. | | Deposit date: | 2017-09-19 | | Release date: | 2018-09-19 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Crystal structure and activation mechanism of DR3 death domain.
Febs J., 286, 2019
|
|
5ZNY
 
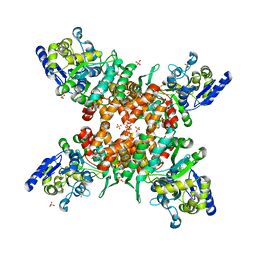 | | Structure of mDR3_DD-C363G with MBP tag | | Descriptor: | Maltose-binding periplasmic protein,Tumor necrosis factor receptor superfamily, member 25, SULFATE ION | | Authors: | Yin, X, Jin, T. | | Deposit date: | 2018-04-11 | | Release date: | 2019-04-17 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.74 Å) | | Cite: | Crystal structure and activation mechanism of DR3 death domain.
Febs J., 286, 2019
|
|
8HGI
 
 | | Crystal structure of VNAR aGFP14 in complex with GFP | | Descriptor: | GFP, VNAR aGFP14 | | Authors: | Zheng, P, Zhu, C, Jin, T. | | Deposit date: | 2022-11-14 | | Release date: | 2023-09-27 | | Last modified: | 2024-10-02 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | Selection, identification and crystal structure of shark-derived single-domain antibodies against a green fluorescent protein.
Int.J.Biol.Macromol., 247, 2023
|
|
7FCP
 
 | |
7FCQ
 
 | |
7F4W
 
 | | Complex structure of HLA2402 with recognizing SARS-CoV-2 epitope pep4 | | Descriptor: | Beta-2-microglobulin, MHC class I antigen, SARS-CoV-2 T-cell Epitope pep4 | | Authors: | Deng, S, Jin, T. | | Deposit date: | 2021-06-21 | | Release date: | 2021-08-25 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (2.9 Å) | | Cite: | Profiling CD8 + T cell epitopes of COVID-19 convalescents reveals reduced cellular immune responses to SARS-CoV-2 variants.
Cell Rep, 36, 2021
|
|
7EU2
 
 | | Complex structure of HLA0201 with recognizing SARS-CoV-2 epitope S1 | | Descriptor: | Beta-2-microglobulin, MHC class I antigen, SARS-CoV-2 T-cell Epitope S1 | | Authors: | Deng, S, Jin, T. | | Deposit date: | 2021-05-15 | | Release date: | 2021-08-25 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Profiling CD8 + T cell epitopes of COVID-19 convalescents reveals reduced cellular immune responses to SARS-CoV-2 variants.
Cell Rep, 36, 2021
|
|
7VZO
 
 | |
7WBO
 
 | | Crystal structure of Sarcoplasmic Calcium-Binding Protein from Scylla paramamosain | | Descriptor: | 1,2-ETHANEDIOL, CALCIUM ION, SODIUM ION, ... | | Authors: | Chen, Y, Jin, T, Liu, G. | | Deposit date: | 2021-12-17 | | Release date: | 2022-12-21 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Crystal Structure Analysis of Sarcoplasmic-Calcium-Binding Protein: An Allergen in Scylla paramamosain.
J.Agric.Food Chem., 71, 2023
|
|
5WQR
 
 | | High resolution structure of high-potential iron-sulfur protein in the reduced state | | Descriptor: | GLYCEROL, High-potential iron-sulfur protein, IRON/SULFUR CLUSTER, ... | | Authors: | Ohno, H, Takeda, K, Niwa, S, Tsujinaka, T, Hanazono, Y, Hirano, Y, Miki, K. | | Deposit date: | 2016-11-28 | | Release date: | 2017-06-07 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (0.8 Å) | | Cite: | Crystallographic characterization of the high-potential iron-sulfur protein in the oxidized state at 0.8 angstrom resolution
PLoS ONE, 12, 2017
|
|
5WQQ
 
 | | High resolution structure of high-potential iron-sulfur protein in the oxidized state | | Descriptor: | GLYCEROL, High-potential iron-sulfur protein, IRON/SULFUR CLUSTER, ... | | Authors: | Ohno, H, Takeda, K, Niwa, S, Tsujinaka, T, Hanazono, Y, Hirano, Y, Miki, K. | | Deposit date: | 2016-11-28 | | Release date: | 2017-06-07 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (0.8 Å) | | Cite: | Crystallographic characterization of the high-potential iron-sulfur protein in the oxidized state at 0.8 angstrom resolution
PLoS ONE, 12, 2017
|
|
