5M7W
 
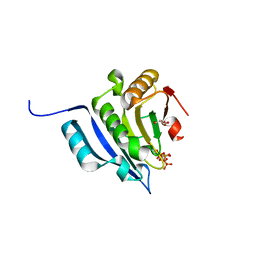 | | Translation initiation factor 4E in complex with (SP)-m2(7,2'O)GppSpG mRNA 5' cap analog (beta-S-ARCA D2) | | Descriptor: | Eukaryotic translation initiation factor 4E, GLYCEROL, [(2~{R},3~{R},4~{R},5~{R})-5-(2-azanyl-7-methyl-6-oxidanylidene-1~{H}-purin-7-ium-9-yl)-4-methoxy-3-oxidanyl-oxolan-2-yl]methyl [phosphonooxy(sulfanyl)phosphoryl] hydrogen phosphate | | Authors: | Warminski, M, Nowak, E, Kowalska, J, Jemielity, J, Nowotny, M. | | Deposit date: | 2016-10-28 | | Release date: | 2017-12-20 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (1.97 Å) | | Cite: | Translation initiation factor 4E in complex with (SP)-m2(7,2'O)GppSpG mRNA 5' cap analog (beta-S-ARCA D2)
To Be Published
|
|
5M81
 
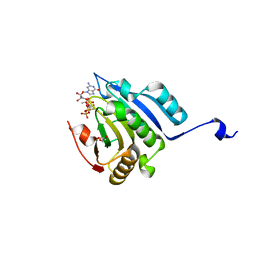 | | Translation initiation factor 4E in complex with (SP)-iPr-m7GppSpG mRNA 5' cap analog | | Descriptor: | Eukaryotic translation initiation factor 4E, GLYCEROL, [[[(3~{a}~{R},4~{R},6~{R},6~{a}~{R})-4-(2-azanyl-7-methyl-6-oxidanylidene-1~{H}-purin-7-ium-9-yl)-2,2-dimethyl-3~{a},4,6,6~{a}-tetrahydrofuro[3,4-d][1,3]dioxol-6-yl]methoxy-oxidanyl-phosphoryl]oxy-sulfanyl-phosphoryl] [(2~{R},3~{S},4~{R},5~{R})-5-(2-azanyl-6-oxidanylidene-3~{H}-purin-9-yl)-3,4-bis(oxidanyl)oxolan-2-yl]methyl hydrogen phosphate | | Authors: | Warminski, M, Nowak, E, Kowalska, J, Jemielity, J, Nowotny, M. | | Deposit date: | 2016-10-28 | | Release date: | 2017-12-20 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Translation initiation factor 4E in complex with (SP)-iPr-m7GppSpG mRNA 5' cap analog
To Be Published
|
|
7T7H
 
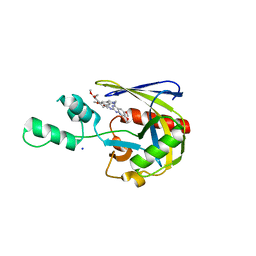 | | Crystal structure of Vaccinia Virus decapping enzyme D9 in complex with inhibitor CP100356 | | Descriptor: | 4-(6,7-dimethoxy-3,4-dihydroisoquinolin-2(1H)-yl)-N-[2-(3,4-dimethoxyphenyl)ethyl]-6,7-dimethoxyquinazolin-2-amine, DNA repair NTP-phosphohydrolase, SODIUM ION | | Authors: | Peters, J.K, Gross, J.D. | | Deposit date: | 2021-12-15 | | Release date: | 2022-06-15 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (1.78000259 Å) | | Cite: | Fluorescence-Based Activity Screening Assay Reveals Small Molecule Inhibitors of Vaccinia Virus mRNA Decapping Enzyme D9.
Acs Chem.Biol., 17, 2022
|
|
2JVB
 
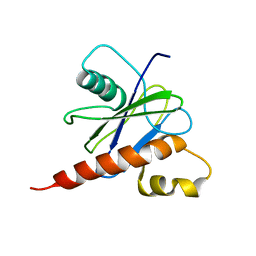 | |
6AM0
 
 | |
