3IHG
 
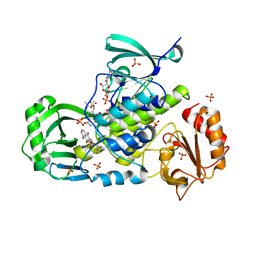 | | Crystal structure of a ternary complex of aklavinone-11 hydroxylase with FAD and aklavinone | | Descriptor: | FLAVIN-ADENINE DINUCLEOTIDE, RdmE, SULFATE ION, ... | | Authors: | Lindqvist, Y, Koskiniemi, H, Jansson, A, Sandalova, T, Schneider, G. | | Deposit date: | 2009-07-30 | | Release date: | 2009-09-29 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (2.49 Å) | | Cite: | Structural basis for substrate recognition and specificity in aklavinone-11-hydroxylase from rhodomycin biosynthesis.
J.Mol.Biol., 393, 2009
|
|
2C95
 
 | | Structure of adenylate kinase 1 in complex with P1,P4-di(adenosine) tetraphosphate | | Descriptor: | ADENYLATE KINASE 1, BIS(ADENOSINE)-5'-TETRAPHOSPHATE, MALONATE ION | | Authors: | Bunkoczi, G, Filippakopoulos, P, Jansson, A, Longman, E, von Delft, F, Edwards, A, Arrowsmith, C, Sundstrom, M, Knapp, S. | | Deposit date: | 2005-12-09 | | Release date: | 2007-02-20 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (1.71 Å) | | Cite: | Structure of Adenylate Kinase 1 in Complex with P1, P4-Di(Adenosine)Tetraphosphate
To be Published
|
|
2BWJ
 
 | | Structure of adenylate kinase 5 | | Descriptor: | ADENOSINE MONOPHOSPHATE, ADENYLATE KINASE 5, CHLORIDE ION, ... | | Authors: | Bunkoczi, G, Filippakopoulos, P, Fedorov, O, Jansson, A, Longman, E, Ugochukwu, E, Knapp, S, von Delft, F, Arrowsmith, C, Edwards, A, Sundstrom, M, Weigelt, J. | | Deposit date: | 2005-07-14 | | Release date: | 2005-07-21 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Structure of Adenylate Kinase 5
To be Published
|
|
5LC2
 
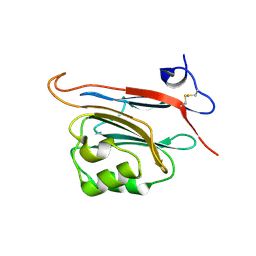 | | Xray structure of human FAM3C ILEI monomer | | Descriptor: | Protein FAM3C | | Authors: | Johansson, P, Jansson, A. | | Deposit date: | 2016-06-19 | | Release date: | 2017-08-02 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | The interleukin-like epithelial-mesenchymal transition inducer ILEI exhibits a non-interleukin-like fold and is active as a domain-swapped dimer.
J. Biol. Chem., 292, 2017
|
|
5LC3
 
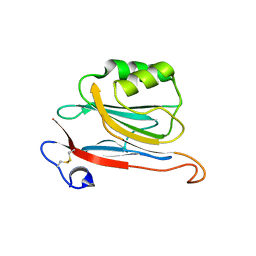 | | Xray structure of mouse FAM3C ILEI monomer | | Descriptor: | Protein FAM3C | | Authors: | Johansson, P, Jansson, A. | | Deposit date: | 2016-06-19 | | Release date: | 2017-08-02 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | The interleukin-like epithelial-mesenchymal transition inducer ILEI exhibits a non-interleukin-like fold and is active as a domain-swapped dimer.
J. Biol. Chem., 292, 2017
|
|
5LC4
 
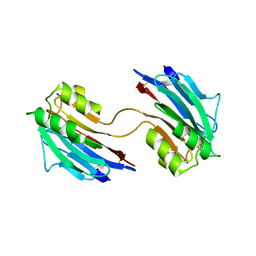 | | Xray structure of mouse FAM3C ILEI dimer | | Descriptor: | Protein FAM3C | | Authors: | Johansson, P, Jansson, A. | | Deposit date: | 2016-06-19 | | Release date: | 2017-08-02 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (1.84 Å) | | Cite: | The interleukin-like epithelial-mesenchymal transition inducer ILEI exhibits a non-interleukin-like fold and is active as a domain-swapped dimer.
J. Biol. Chem., 292, 2017
|
|
1ZV4
 
 | | Structure of the Regulator of G-Protein Signaling 17 (RGSZ2) | | Descriptor: | Regulator of G-protein signaling 17 | | Authors: | Schoch, G.A, Jansson, A, Elkins, J.M, Haroniti, A, Niesen, F.H, Bunkoczi, G, Lee, W.H, Turnbull, A.P, Yang, X, Sundstrom, M, Arrowsmith, C, Edwards, A, Marsden, B, Gileadi, O, Ball, L, von Delft, F, Doyle, D.A, Structural Genomics Consortium (SGC) | | Deposit date: | 2005-06-01 | | Release date: | 2005-06-28 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Structural diversity in the RGS domain and its interaction with heterotrimeric G protein alpha-subunits.
Proc.Natl.Acad.Sci.Usa, 105, 2008
|
|
6S4U
 
 | | LXRbeta ligand binding domain in comlpex with small molecule inhibitors | | Descriptor: | 6-[4-[[3-oxidanyl-1,1-bis(oxidanylidene)-5-phenyl-2-propan-2-yl-3~{H}-1,2-thiazol-4-yl]amino]butyl]pyridine-2-sulfonamide, Oxysterols receptor LXR-beta | | Authors: | Sandmark, J, Jansson, A. | | Deposit date: | 2019-06-28 | | Release date: | 2019-11-27 | | Last modified: | 2024-05-15 | | Method: | X-RAY DIFFRACTION (2.81 Å) | | Cite: | Structural analysis identifies an escape route from the adverse lipogenic effects of liver X receptor ligands.
Commun Biol, 2, 2019
|
|
6S4T
 
 | | LXRbeta ligand binding domain in comlpex with small molecule inhibitors | | Descriptor: | 2-[4-[[3-[3-(phenylmethyl)-8-(trifluoromethyl)quinolin-4-yl]phenoxy]methyl]phenyl]ethanoic acid, Oxysterols receptor LXR-beta | | Authors: | Sandmark, J, Jansson, A. | | Deposit date: | 2019-06-28 | | Release date: | 2019-11-27 | | Last modified: | 2024-05-15 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Structural analysis identifies an escape route from the adverse lipogenic effects of liver X receptor ligands.
Commun Biol, 2, 2019
|
|
6S4N
 
 | | LXRbeta ligand binding domain in comlpex with small molecule inhibitors | | Descriptor: | 2-[5-chloranyl-6-[4-[[1,1,3-tris(oxidanylidene)-5-phenyl-2-propan-2-yl-1,2-thiazol-4-yl]amino]piperidin-1-yl]pyridin-3-yl]ethanoic acid, Oxysterols receptor LXR-beta, SULFATE ION | | Authors: | Sandmark, J, Jansson, A. | | Deposit date: | 2019-06-28 | | Release date: | 2019-11-27 | | Last modified: | 2024-05-15 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Structural analysis identifies an escape route from the adverse lipogenic effects of liver X receptor ligands.
Commun Biol, 2, 2019
|
|
