2GC2
 
 | |
1H7Y
 
 | | Translationally Controlled Tumor-associated Protein p23fyp from Schizosaccharomyces pombe | | Descriptor: | TRANSLATIONALLY CONTROLLED TUMOR PROTEIN | | Authors: | Thaw, P, Baxter, N.J, Sedelnikova, S.E, Price, C, Waltho, J.P, Craven, C.J. | | Deposit date: | 2001-01-19 | | Release date: | 2001-08-09 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | Structure of TCTP reveals unexpected relationship with guanine nucleotide-free chaperones.
Nat. Struct. Biol., 8, 2001
|
|
1H6Q
 
 | | Translationally Controlled Tumor-associated Protein p23fyp from Schizosaccharomyces pombe | | Descriptor: | TRANSLATIONALLY CONTROLLED TUMOR PROTEIN | | Authors: | Thaw, P, Baxter, N.J, Sedelnikova, S.E, Price, C, Waltho, J.P, Craven, C.J. | | Deposit date: | 2001-06-20 | | Release date: | 2001-08-09 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | Structure of TCTP reveals unexpected relationship with guanine nucleotide-free chaperones.
Nat. Struct. Biol., 8, 2001
|
|
2MKX
 
 | |
6RK4
 
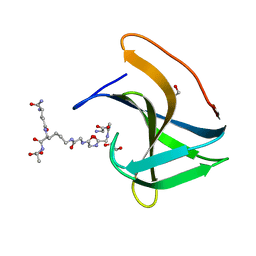 | | Lysostaphin SH3b P4-G5 complex, synchrotron dataset | | Descriptor: | (2~{R})-2-[[(2~{S})-2-[[(4~{R})-5-azanyl-4-[[(2~{S})-2-azanylpropanoyl]amino]-5-oxidanylidene-pentanoyl]amino]-6-[2-[2-[2-[2-(2-azanylethanoylamino)ethanoylamino]ethanoylamino]ethanoylamino]ethanoylamino]hexanoyl]amino]propanoic acid, 1,2-ETHANEDIOL, Lysostaphin | | Authors: | Walters-Morgan, H, Lovering, A.L. | | Deposit date: | 2019-04-30 | | Release date: | 2019-10-16 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.43 Å) | | Cite: | Two-site recognition of Staphylococcus aureus peptidoglycan by lysostaphin SH3b.
Nat.Chem.Biol., 16, 2020
|
|
6RJE
 
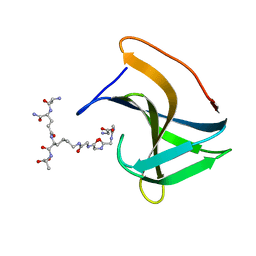 | | Lysostaphin SH3b P4-G5 complex, homesource dataset | | Descriptor: | (2~{R})-2-[[(2~{S})-2-[[(4~{R})-5-azanyl-4-[[(2~{S})-2-azanylpropanoyl]amino]-5-oxidanylidene-pentanoyl]amino]-6-[2-[2-[2-[2-(2-azanylethanoylamino)ethanoylamino]ethanoylamino]ethanoylamino]ethanoylamino]hexanoyl]amino]propanoic acid, Lysostaphin | | Authors: | Walters-Morgan, H, Lovering, A.L. | | Deposit date: | 2019-04-26 | | Release date: | 2019-10-16 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Two-site recognition of Staphylococcus aureus peptidoglycan by lysostaphin SH3b.
Nat.Chem.Biol., 16, 2020
|
|
6SUZ
 
 | |
6SV2
 
 | |
5O6R
 
 | |
5OJZ
 
 | |
5OK0
 
 | | Structure of the D10N mutant of beta-phosphoglucomutase from Lactococcus lactis trapped with native reaction intermediate beta-glucose 1,6-bisphosphate to 2.2A resolution. | | Descriptor: | 1,3-PROPANDIOL, 1,6-di-O-phosphono-beta-D-glucopyranose, Beta-phosphoglucomutase, ... | | Authors: | Robertson, A.J, Bisson, C. | | Deposit date: | 2017-07-25 | | Release date: | 2018-08-29 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (2.15 Å) | | Cite: | van der Waals Contact between Nucleophile and Transferring Phosphorus Is Insufficient To Achieve Enzyme Transition-State Architecture
Acs Catalysis, 2018
|
|
5OK2
 
 | |
5O6P
 
 | |
5OK1
 
 | |
2WF7
 
 | | Structure of Beta-Phosphoglucomutase inhibited with Glucose-6- phosphonate and Aluminium tetrafluoride | | Descriptor: | 6,7-dideoxy-7-phosphono-beta-D-gluco-heptopyranose, BETA-PHOSPHOGLUCOMUTASE, MAGNESIUM ION, ... | | Authors: | Bowler, M.W, Baxter, N.J, Webster, C.E, Pollard, S, Alizadeh, T, Hounslow, A.M, Cliff, M.J, Bermel, W, Williams, N.H, Hollfelder, F, Blackburn, G.M, Waltho, J.P. | | Deposit date: | 2009-04-03 | | Release date: | 2010-05-19 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (1.05 Å) | | Cite: | Alpha-Fluorophosphonates Reveal How a Phosphomutase Conserves Transition State Conformation Over Hexose Recognition in its Two-Step Reaction.
Proc.Natl.Acad.Sci.USA, 111, 2014
|
|
